Lentivirus Manufacturing Process for Primary T-Cell Biofactory Production
- PMID: 32390316
- PMCID: PMC7526617
- DOI: 10.1002/adbi.201900288
Lentivirus Manufacturing Process for Primary T-Cell Biofactory Production
Abstract
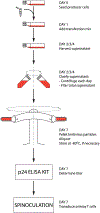
A process for maximizing the titer of lentivirus particles, deemed to be a necessity for transducing primary cells, is developed. Lentivirus particles, with a set of transgenes encoding an artificial cell-signaling pathway, are used to transform primary T cells as vectors for calibrated synthesis of desired proteins in situ, that is, T-cell biofactory cells. The process is also used to generate primary T cells expressing antigen-specific chimeric antigen receptors, that is, CAR T cells. The two differently engineered primary T cells are expanded and validated for their respective functions, that is, calibrated synthesis of desired proteins upon engaging the target cells, which is specific for the T-cell biofactory cells, and cytolysis of the target cells common to both types of cells. The process is compliant with current Good Manufacturing Practices and can be used to support the scale-up for clinical translation.
Keywords: CAR T cells; cell engineering; cell manufacturing; drug delivery; high-titer lentivirus production.
© 2020 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Figures


Similar articles
-
Engineered Ovarian Cancer Cell Lines for Validation of CAR T Cell Function.Adv Biosyst. 2020 Jan;4(1):e1900224. doi: 10.1002/adbi.201900224. Epub 2019 Nov 29. Adv Biosyst. 2020. PMID: 32293122 Free PMC article.
-
Comparative analysis and optimization of protocols for producing recombinant lentivirus carrying the anti-Her2 chimeric antigen receptor gene.J Gene Med. 2018 Jul;20(7-8):e3027. doi: 10.1002/jgm.3027. Epub 2018 Jul 6. J Gene Med. 2018. PMID: 29851200
-
Optimized Production of Lentiviral Vectors for CAR-T Cell.Methods Mol Biol. 2020;2086:69-76. doi: 10.1007/978-1-0716-0146-4_5. Methods Mol Biol. 2020. PMID: 31707668
-
Production of CAR T-cells by GMP-grade lentiviral vectors: latest advances and future prospects.Crit Rev Clin Lab Sci. 2019 Sep;56(6):393-419. doi: 10.1080/10408363.2019.1633512. Epub 2019 Jul 17. Crit Rev Clin Lab Sci. 2019. PMID: 31314617 Review.
-
Next generation chimeric antigen receptor T cells: safety strategies to overcome toxicity.Mol Cancer. 2019 Aug 20;18(1):125. doi: 10.1186/s12943-019-1057-4. Mol Cancer. 2019. PMID: 31429760 Free PMC article. Review.
Cited by
-
Primary T-cell-based delivery platform for in vivo synthesis of engineered proteins.Bioeng Transl Med. 2023 Oct 7;9(1):e10605. doi: 10.1002/btm2.10605. eCollection 2024 Jan. Bioeng Transl Med. 2023. PMID: 38193126 Free PMC article.
-
Electrically regulated cell-based intervention for viral infections.Bioeng Transl Med. 2022 Nov 15;8(2):e10434. doi: 10.1002/btm2.10434. eCollection 2023 Mar. Bioeng Transl Med. 2022. PMID: 36925710 Free PMC article.
-
NK-Cell Biofactory as an Off-the-Shelf Cell-based Vector for Targeted In Situ Synthesis of Engineered Proteins.Adv Biol (Weinh). 2021 Jul;5(7):e2000298. doi: 10.1002/adbi.202000298. Epub 2021 Apr 19. Adv Biol (Weinh). 2021. PMID: 33871182 Free PMC article.
-
Genetically engineered pair of cells for serological testing and its application for SARS-CoV-2.Bioeng Transl Med. 2023 Mar 24;8(3):e10508. doi: 10.1002/btm2.10508. eCollection 2023 May. Bioeng Transl Med. 2023. PMID: 37206248 Free PMC article.
-
Optimized conditions for gene transduction into primary immune cells using viral vectors.Sci Rep. 2023 Jul 31;13(1):12365. doi: 10.1038/s41598-023-39597-2. Sci Rep. 2023. PMID: 37524755 Free PMC article.
References
-
- Ornstein MC & Rini BI (2016) Pharmacokinetically-guided dosing of oral drugs: true precision oncology? Clinical Cancer Research:clincanres.1833.2016. - PubMed
-
- Schmidinger M, Danesi R, Jones R, McDermott R, Pyle L, Rini B, & Négrier S (2018) Individualized dosing with axitinib: rationale and practical guidance. Future Oncology 14(9):861–875. - PubMed
-
- Mathijssen RHJ, de Jong FA, Loos WJ, van der Bol JM, Verweij J, & Sparreboom A (2007) Flat-Fixed Dosing Versus Body Surface Area–Based Dosing of Anticancer Drugs in Adults: Does It Make a Difference? The Oncologist 12(8):913–923. - PubMed
-
- Deeken JF, Figg WD, Bates SE, & Sparreboom A (2007) Toward individualized treatment: prediction of anticancer drug disposition and toxicity with pharmacogenetics. Anti-Cancer Drugs 18(2):111–126. - PubMed
-
- Baker SD, Verweij J, Rowinsky EK, Donehower RC, Schellens JHM, Grochow LB, & Sparreboom A (2002) Role of Body Surface Area in Dosing of Investigational Anticancer Agents in Adults, 1991–2001. JNCI: Journal of the National Cancer Institute 94(24):1883–1888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

