Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies
- PMID: 32375025
- PMCID: PMC7199733
- DOI: 10.1016/j.cell.2020.04.031
Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies
Erratum in
-
Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies.Cell. 2020 Jun 11;181(6):1436-1441. doi: 10.1016/j.cell.2020.05.047. Cell. 2020. PMID: 32531248 Free PMC article. No abstract available.
Abstract
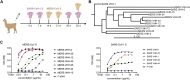
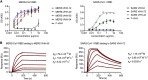
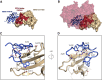
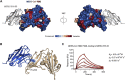
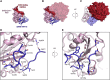
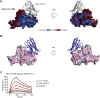
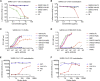
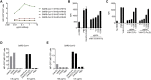
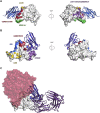
Coronaviruses make use of a large envelope protein called spike (S) to engage host cell receptors and catalyze membrane fusion. Because of the vital role that these S proteins play, they represent a vulnerable target for the development of therapeutics. Here, we describe the isolation of single-domain antibodies (VHHs) from a llama immunized with prefusion-stabilized coronavirus spikes. These VHHs neutralize MERS-CoV or SARS-CoV-1 S pseudotyped viruses, respectively. Crystal structures of these VHHs bound to their respective viral targets reveal two distinct epitopes, but both VHHs interfere with receptor binding. We also show cross-reactivity between the SARS-CoV-1 S-directed VHH and SARS-CoV-2 S and demonstrate that this cross-reactive VHH neutralizes SARS-CoV-2 S pseudotyped viruses as a bivalent human IgG Fc-fusion. These data provide a molecular basis for the neutralization of pathogenic betacoronaviruses by VHHs and suggest that these molecules may serve as useful therapeutics during coronavirus outbreaks.
Keywords: COVID-19; MERS; SARS; SARS-CoV-2; nanobody.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests K.S.C., N.W., B.S.G., and J.S.M. are inventors on US patent application no. 62/412,703, entitled “Prefusion Coronavirus Spike Proteins and Their Use.” D.W., K.S.C., N.W., B.S.G., and J.S.M. are inventors on US patent application no. 62/972,886, entitled “2019-nCoV Vaccine.” D.W., D.D.V., B.S.G., B.S., X.S., and J.S.M. are inventors on US patent application no. 62/988,610, entitled “Coronavirus Binders.” D.W., L.v.S., N.C., B.S., X.S., and J.S.M. are inventors on US patent application no. 62/991,408, entitled “SARS-CoV-2 Virus Binders.”
Figures


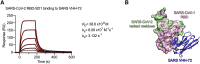


Comment in
-
Finding Camel-ot: A Holy Grail against pandemic SARS-CoV-2?Sci Immunol. 2020 Jul 3;5(49):eabd4758. doi: 10.1126/sciimmunol.abd4758. Sci Immunol. 2020. PMID: 32620562 Free PMC article.
Similar articles
-
Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants.Nature. 2021 Jul;595(7866):278-282. doi: 10.1038/s41586-021-03676-z. Epub 2021 Jun 7. Nature. 2021. PMID: 34098567 Free PMC article.
-
An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction.Nat Commun. 2020 Sep 4;11(1):4420. doi: 10.1038/s41467-020-18174-5. Nat Commun. 2020. PMID: 32887876 Free PMC article.
-
Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.Nature. 2020 Jul;583(7815):290-295. doi: 10.1038/s41586-020-2349-y. Epub 2020 May 18. Nature. 2020. PMID: 32422645
-
Therapeutic nanobodies against SARS-CoV-2 and other pathogenic human coronaviruses.J Nanobiotechnology. 2024 May 31;22(1):304. doi: 10.1186/s12951-024-02573-7. J Nanobiotechnology. 2024. PMID: 38822339 Free PMC article. Review.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
Cited by
-
An intracellular nanobody targeting T4SS effector inhibits Ehrlichia infection.Proc Natl Acad Sci U S A. 2021 May 4;118(18):e2024102118. doi: 10.1073/pnas.2024102118. Proc Natl Acad Sci U S A. 2021. PMID: 33903242 Free PMC article.
-
Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies.Biochem Biophys Res Commun. 2021 Jan 29;538:192-203. doi: 10.1016/j.bbrc.2020.10.012. Epub 2020 Oct 10. Biochem Biophys Res Commun. 2021. PMID: 33069360 Free PMC article. Review.
-
PCRCR complex is essential for invasion of human erythrocytes by Plasmodium falciparum.Nat Microbiol. 2022 Dec;7(12):2039-2053. doi: 10.1038/s41564-022-01261-2. Epub 2022 Nov 17. Nat Microbiol. 2022. PMID: 36396942 Free PMC article.
-
UCSF ChimeraX: Structure visualization for researchers, educators, and developers.Protein Sci. 2021 Jan;30(1):70-82. doi: 10.1002/pro.3943. Epub 2020 Oct 22. Protein Sci. 2021. PMID: 32881101 Free PMC article.
-
Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.SSRN [Preprint]. 2020 Jul 21:3639618. doi: 10.2139/ssrn.3639618. SSRN. 2020. Update in: Cell Rep. 2020 Oct 27;33(4):108322. doi: 10.1016/j.celrep.2020.108322 PMID: 32742241 Free PMC article. Updated. Preprint.
References
-
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

