Too Much of a Good Thing: How Ectopic DNA Replication Affects Bacterial Replication Dynamics
- PMID: 32351461
- PMCID: PMC7174701
- DOI: 10.3389/fmicb.2020.00534
Too Much of a Good Thing: How Ectopic DNA Replication Affects Bacterial Replication Dynamics
Abstract
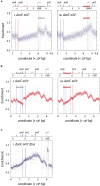
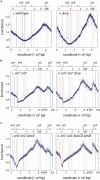
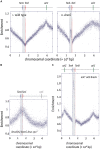
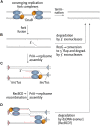
Each cell division requires the complete and accurate duplication of the entire genome. In bacteria, the duplication process of the often-circular chromosomes is initiated at a single origin per chromosome, resulting in two replication forks that traverse the chromosome in opposite directions. DNA synthesis is completed once the two forks fuse in a region diametrically opposite the origin. In some bacteria, such as Escherichia coli, the region where forks fuse forms a specialized termination area. Polar replication fork pause sites flanking this area can pause the progression of replication forks, thereby allowing forks to enter but not to leave. Transcription of all required genes has to take place simultaneously with genome duplication. As both of these genome trafficking processes share the same template, conflicts are unavoidable. In this review, we focus on recent attempts to add additional origins into various ectopic chromosomal locations of the E. coli chromosome. As ectopic origins disturb the native replichore arrangements, the problems resulting from such perturbations can give important insights into how genome trafficking processes are coordinated and the problems that arise if this coordination is disturbed. The data from these studies highlight that head-on replication-transcription conflicts are indeed highly problematic and multiple repair pathways are required to restart replication forks arrested at obstacles. In addition, the existing data also demonstrate that the replication fork trap in E. coli imposes significant constraints to genome duplication if ectopic origins are active. We describe the current models of how replication fork fusion events can cause serious problems for genome duplication, as well as models of how such problems might be alleviated both by a number of repair pathways as well as the replication fork trap system. Considering the problems associated both with head-on replication-transcription conflicts as well as head-on replication fork fusion events might provide clues of how these genome trafficking issues have contributed to shape the distinct architecture of bacterial chromosomes.
Keywords: 3′ exonuclease; bacterial replication dynamics; ectopic replication origins; recG gene; replication; termination of DNA replication; transcription.
Copyright © 2020 Syeda, Dimude, Skovgaard and Rudolph.
Figures


Similar articles
-
Interplay between chromosomal architecture and termination of DNA replication in bacteria.Front Microbiol. 2023 Jun 26;14:1180848. doi: 10.3389/fmicb.2023.1180848. eCollection 2023. Front Microbiol. 2023. PMID: 37434703 Free PMC article. Review.
-
Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome.Genes (Basel). 2018 Jul 27;9(8):376. doi: 10.3390/genes9080376. Genes (Basel). 2018. PMID: 30060465 Free PMC article.
-
Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli.Genes (Basel). 2016 Jul 25;7(8):40. doi: 10.3390/genes7080040. Genes (Basel). 2016. PMID: 27463728 Free PMC article. Review.
-
The Consequences of Replicating in the Wrong Orientation: Bacterial Chromosome Duplication without an Active Replication Origin.mBio. 2015 Nov 3;6(6):e01294-15. doi: 10.1128/mBio.01294-15. mBio. 2015. PMID: 26530381 Free PMC article.
-
Chromosomal over-replication in Escherichia coli recG cells is triggered by replication fork fusion and amplified if replichore symmetry is disturbed.Nucleic Acids Res. 2018 Sep 6;46(15):7701-7715. doi: 10.1093/nar/gky566. Nucleic Acids Res. 2018. PMID: 29982635 Free PMC article.
Cited by
-
Controlling Genome Topology with Sequences that Trigger Post-replication Gap Formation During Replisome Passage: The E. coli RRS Elements.bioRxiv [Preprint]. 2024 Apr 10:2023.10.01.560376. doi: 10.1101/2023.10.01.560376. bioRxiv. 2024. Update in: Nucleic Acids Res. 2024 Jun 24;52(11):6392-6405. doi: 10.1093/nar/gkae320 PMID: 37873128 Free PMC article. Updated. Preprint.
-
Single-nucleotide resolution detection of Topo IV cleavage activity in the Escherichia coli genome with Topo-Seq.Front Microbiol. 2023 Apr 6;14:1160736. doi: 10.3389/fmicb.2023.1160736. eCollection 2023. Front Microbiol. 2023. PMID: 37089538 Free PMC article.
-
Subtelomeres are fast-evolving regions of the Streptomyces linear chromosome.Microb Genom. 2019 Sep;7(6):000525. doi: 10.1099/mgen.0.000525. Microb Genom. 2019. PMID: 33749576 Free PMC article.
-
A Fork Trap in the Chromosomal Termination Area Is Highly Conserved across All Escherichia coli Phylogenetic Groups.Int J Mol Sci. 2021 Jul 25;22(15):7928. doi: 10.3390/ijms22157928. Int J Mol Sci. 2021. PMID: 34360694 Free PMC article.
-
Interplay between chromosomal architecture and termination of DNA replication in bacteria.Front Microbiol. 2023 Jun 26;14:1180848. doi: 10.3389/fmicb.2023.1180848. eCollection 2023. Front Microbiol. 2023. PMID: 37434703 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

