Division of labour in a matrix, rather than phagocytosis or endosymbiosis, as a route for the origin of eukaryotic cells
- PMID: 32345370
- PMCID: PMC7187495
- DOI: 10.1186/s13062-020-00260-9
Division of labour in a matrix, rather than phagocytosis or endosymbiosis, as a route for the origin of eukaryotic cells
Abstract
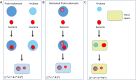
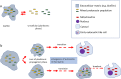
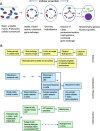
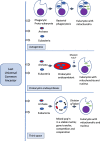
Two apparently irreconcilable models dominate research into the origin of eukaryotes. In one model, amitochondrial proto-eukaryotes emerged autogenously from the last universal common ancestor of all cells. Proto-eukaryotes subsequently acquired mitochondrial progenitors by the phagocytic capture of bacteria. In the second model, two prokaryotes, probably an archaeon and a bacterial cell, engaged in prokaryotic endosymbiosis, with the species resident within the host becoming the mitochondrial progenitor. Both models have limitations. A search was therefore undertaken for alternative routes towards the origin of eukaryotic cells. The question was addressed by considering classes of potential pathways from prokaryotic to eukaryotic cells based on considerations of cellular topology. Among the solutions identified, one, called here the "third-space model", has not been widely explored. A version is presented in which an extracellular space (the third-space), serves as a proxy cytoplasm for mixed populations of archaea and bacteria to "merge" as a transitionary complex without obligatory endosymbiosis or phagocytosis and to form a precursor cell. Incipient nuclei and mitochondria diverge by division of labour. The third-space model can accommodate the reorganization of prokaryote-like genomes to a more eukaryote-like genome structure. Nuclei with multiple chromosomes and mitosis emerge as a natural feature of the model. The model is compatible with the loss of archaeal lipid biochemistry while retaining archaeal genes and provides a route for the development of membranous organelles such as the Golgi apparatus and endoplasmic reticulum. Advantages, limitations and variations of the "third-space" models are discussed. REVIEWERS: This article was reviewed by Damien Devos, Buzz Baum and Michael Gray.
Keywords: Archaea; Bacteria; Biofilm; Chromosomes; Eukaryogenesis; Eukaryotes; Evolution; Hypothesis; Matrix; Membranes; Mitochondria; Prokaryotes.
Conflict of interest statement
The author declares that he has no competing interests.
Figures
Similar articles
-
The Physiology of Phagocytosis in the Context of Mitochondrial Origin.Microbiol Mol Biol Rev. 2017 Jun 14;81(3):e00008-17. doi: 10.1128/MMBR.00008-17. Print 2017 Sep. Microbiol Mol Biol Rev. 2017. PMID: 28615286 Free PMC article. Review.
-
Energetics and genetics across the prokaryote-eukaryote divide.Biol Direct. 2011 Jun 30;6:35. doi: 10.1186/1745-6150-6-35. Biol Direct. 2011. PMID: 21714941 Free PMC article.
-
Endosymbiosis before eukaryotes: mitochondrial establishment in protoeukaryotes.Cell Mol Life Sci. 2020 Sep;77(18):3503-3523. doi: 10.1007/s00018-020-03462-6. Epub 2020 Feb 1. Cell Mol Life Sci. 2020. PMID: 32008087 Free PMC article. Review.
-
Mitochondria, the Cell Cycle, and the Origin of Sex via a Syncytial Eukaryote Common Ancestor.Genome Biol Evol. 2016 Jul 2;8(6):1950-70. doi: 10.1093/gbe/evw136. Genome Biol Evol. 2016. PMID: 27345956 Free PMC article.
-
Was the Mitochondrion Necessary to Start Eukaryogenesis?Trends Microbiol. 2019 Feb;27(2):96-104. doi: 10.1016/j.tim.2018.10.005. Epub 2018 Nov 19. Trends Microbiol. 2019. PMID: 30466901 Review.
Cited by
-
Thriving in Oxygen While Preventing ROS Overproduction: No Two Systems Are Created Equal.Front Physiol. 2022 Apr 4;13:874321. doi: 10.3389/fphys.2022.874321. eCollection 2022. Front Physiol. 2022. PMID: 35444563 Free PMC article. Review.
-
Organelle Genome Variation in the Red Algal Genus Ahnfeltia (Florideophyceae).Front Genet. 2021 Sep 27;12:724734. doi: 10.3389/fgene.2021.724734. eCollection 2021. Front Genet. 2021. PMID: 34646303 Free PMC article.
References
-
- Gold DA, Caron A, Fournier GP, Summons RE. Paleoproterozoic sterol biosynthesis and the rise of oxygen. Nature. 2017;543(7645):420–423. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

