Synonymous Dinucleotide Usage: A Codon-Aware Metric for Quantifying Dinucleotide Representation in Viruses
- PMID: 32325924
- PMCID: PMC7232450
- DOI: 10.3390/v12040462
Synonymous Dinucleotide Usage: A Codon-Aware Metric for Quantifying Dinucleotide Representation in Viruses
Abstract
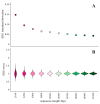
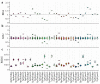
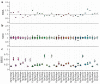
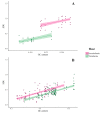
Distinct patterns of dinucleotide representation, such as CpG and UpA suppression, are characteristic of certain viral genomes. Recent research has uncovered vertebrate immune mechanisms that select against specific dinucleotides in targeted viruses. This evidence highlights the importance of systematically examining the dinucleotide composition of viral genomes. We have developed a novel metric, called synonymous dinucleotide usage (SDU), for quantifying dinucleotide representation in coding sequences. Our method compares the abundance of a given dinucleotide to the null hypothesis of equal synonymous codon usage in the sequence. We present a Python3 package, DinuQ, for calculating SDU and other relevant metrics. We have applied this method on two sets of invertebrate- and vertebrate-specific flaviviruses and rhabdoviruses. The SDU shows that the vertebrate viruses exhibit consistently greater under-representation of CpG dinucleotides in all three codon positions in both datasets. In comparison to existing metrics for dinucleotide quantification, the SDU allows for a statistical interpretation of its values by comparing it to a null expectation based on the codon table. Here we apply the method to viruses, but coding sequences of other living organisms can be analysed in the same way.
Keywords: CpG suppression; Flaviviridae; Rhabdoviridae; bioinformatics; dinucleotides; python package; synonymous codon usage.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts.PLoS One. 2009 Jul 20;4(7):e6282. doi: 10.1371/journal.pone.0006282. PLoS One. 2009. PMID: 19617912 Free PMC article.
-
The dinucleotide composition of the Zika virus genome is shaped by conflicting evolutionary pressures in mammalian hosts and mosquito vectors.PLoS Biol. 2021 Apr 19;19(4):e3001201. doi: 10.1371/journal.pbio.3001201. eCollection 2021 Apr. PLoS Biol. 2021. PMID: 33872300 Free PMC article.
-
Causal analysis of CpG suppression in the Mycoplasma genome.Microb Comp Genomics. 2000;5(1):51-8. doi: 10.1089/10906590050145267. Microb Comp Genomics. 2000. PMID: 11011765
-
Does the Zinc Finger Antiviral Protein (ZAP) Shape the Evolution of Herpesvirus Genomes?Viruses. 2021 Sep 17;13(9):1857. doi: 10.3390/v13091857. Viruses. 2021. PMID: 34578438 Free PMC article. Review.
-
Attenuation of Human Respiratory Viruses by Synonymous Genome Recoding.Front Immunol. 2019 Jun 4;10:1250. doi: 10.3389/fimmu.2019.01250. eCollection 2019. Front Immunol. 2019. PMID: 31231383 Free PMC article. Review.
Cited by
-
Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen.PLoS Biol. 2021 Mar 12;19(3):e3001115. doi: 10.1371/journal.pbio.3001115. eCollection 2021 Mar. PLoS Biol. 2021. PMID: 33711012 Free PMC article.
-
The International Virus Bioinformatics Meeting 2022.Viruses. 2022 May 5;14(5):973. doi: 10.3390/v14050973. Viruses. 2022. PMID: 35632715 Free PMC article.
-
Investigating the Diversity and Host Range of Novel Parvoviruses from North American Ducks Using Epidemiology, Phylogenetics, Genome Structure, and Codon Usage Analysis.Viruses. 2021 Jan 28;13(2):193. doi: 10.3390/v13020193. Viruses. 2021. PMID: 33525386 Free PMC article.
-
Codon usage bias and dinucleotide preference in 29 Drosophila species.G3 (Bethesda). 2021 Aug 7;11(8):jkab191. doi: 10.1093/g3journal/jkab191. G3 (Bethesda). 2021. PMID: 34849812 Free PMC article.
-
Significant non-existence of sequences in genomes and proteomes.Nucleic Acids Res. 2021 Apr 6;49(6):3139-3155. doi: 10.1093/nar/gkab139. Nucleic Acids Res. 2021. PMID: 33693858 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

