ZWINT: A potential therapeutic biomarker in patients with glioblastoma correlates with cell proliferation and invasion
- PMID: 32323832
- PMCID: PMC7160549
- DOI: 10.3892/or.2020.7573
ZWINT: A potential therapeutic biomarker in patients with glioblastoma correlates with cell proliferation and invasion
Abstract
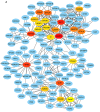
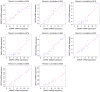
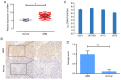
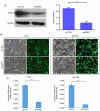
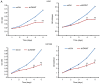
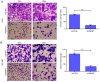
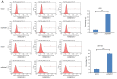
Glioblastoma (GBM) is the most aggressive primary intracranial tumor in adults. Chemoradiotherapy resistance and recurrence after surgery are the main malignant progression factors, leading to a high mortality rate. Therefore, the exploration of novel biomarkers and molecular mechanisms of GBM is urgent. Differentially expressed genes (DEGs) of GBM were screened in a TCGA dataset. Homo sapiens ZW10 interacting kinetochore protein (ZWINT) was found to be upregulated in GBM, which was confirmed by immunohistochemical staining of a tissue microarray. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database. A protein‑protein interaction (PPI) network was established by the STRING database, and hub genes were visualized by Cytoscape. The correlation results were verified with the GSE15824 dataset. Bioinformatic analysis confirmed that ZWINT was significantly positively correlated with kinetochore protein NDC80 homolog (NDC80), serine/threonine‑protein kinase PLK1 (PLK1) and spindle and kinetochore associated complex subunit 1 (SKA1) and together are involved in regulating mitosis and the cell cycle of GBM. ZWINT expression was knocked down in U251 and U87 MG GBM cells by lentiviral vectors carrying a small hairpin RNA (shRNA) targeting ZWINT. The effect of ZWINT silencing on cell proliferation, invasion and apoptosis was determined by the Celigo assay, MTT assay, Transwell assay, flow cytometry and caspase‑3/7 assay in vitro. A subcutaneous xenograft tumor model was established to explore the influence of ZWINT knockdown on GBM growth in vivo. Our preliminary study demonstrated that ZWINT knockdown effectively inhibited proliferation and invasion and induced apoptosis of GBM cells and notably suppressed GBM growth in vivo. Therefore, we speculate that ZWINT may be a potential therapeutic biomarker for GBM, with NDC80 and PLK1 conjointly involved in regulating cell division and the mitotic cell cycle.
Keywords: glioblastoma; bioinformatics; TCGA; GEO; ZWINT; shRNA.
Figures





Similar articles
-
NAMPT: A potential prognostic and therapeutic biomarker in patients with glioblastoma.Oncol Rep. 2019 Sep;42(3):963-972. doi: 10.3892/or.2019.7227. Epub 2019 Jul 10. Oncol Rep. 2019. PMID: 31322259 Free PMC article.
-
PBK as a Potential Biomarker Associated with Prognosis of Glioblastoma.J Mol Neurosci. 2020 Jan;70(1):56-64. doi: 10.1007/s12031-019-01400-1. Epub 2019 Oct 15. J Mol Neurosci. 2020. PMID: 31617063
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
MicroRNAs in human glioblastoma: from bench to beside.Front Biosci (Landmark Ed). 2015 Jan 1;20(1):105-18. doi: 10.2741/4300. Front Biosci (Landmark Ed). 2015. PMID: 25553442 Review.
-
Biologically Active TNIIIA2 Region in Tenascin-C Molecule: A Major Contributor to Elicit Aggressive Malignant Phenotypes From Tumors/Tumor Stroma.Front Immunol. 2020 Dec 9;11:610096. doi: 10.3389/fimmu.2020.610096. eCollection 2020. Front Immunol. 2020. PMID: 33362799 Free PMC article. Review.
Cited by
-
ZWINT is a Promising Therapeutic Biomarker Associated with the Immune Microenvironment of Hepatocellular Carcinoma.Int J Gen Med. 2021 Oct 30;14:7487-7501. doi: 10.2147/IJGM.S340057. eCollection 2021. Int J Gen Med. 2021. PMID: 34744456 Free PMC article.
-
Identification of cervical cancer stem cells using single-cell transcriptomes of normal cervix, cervical premalignant lesions, and cervical cancer.EBioMedicine. 2023 Jun;92:104612. doi: 10.1016/j.ebiom.2023.104612. Epub 2023 May 22. EBioMedicine. 2023. PMID: 37224771 Free PMC article.
-
Bacoside a inhibits the growth of glioma by promoting apoptosis and autophagy in U251 and U87 cells.Naunyn Schmiedebergs Arch Pharmacol. 2024 Apr;397(4):2105-2120. doi: 10.1007/s00210-023-02724-x. Epub 2023 Oct 2. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 37782380
-
Hypoxia-Induced ZWINT Mediates Pancreatic Cancer Proliferation by Interacting With p53/p21.Front Cell Dev Biol. 2021 Nov 24;9:682131. doi: 10.3389/fcell.2021.682131. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34900978 Free PMC article.
-
Unveiling novel prognostic biomarkers and therapeutic targets for HBV-associated hepatocellular carcinoma through integrated bioinformatic analysis.Medicine (Baltimore). 2024 Oct 25;103(43):e40134. doi: 10.1097/MD.0000000000040134. Medicine (Baltimore). 2024. PMID: 39470543 Free PMC article.
References
-
- Biedermann J, Preussler M, Conde M, Peitzsch M, Richter S, Wiedemuth R, Abou-El-Ardat K, Krüger A, Meinhardt M, Schackert G, et al. Mutant IDH1 differently affects redox state and metabolism in glial cells of normal and tumor origin. Cancers (Basel) 2019;11(pii):E2028. doi: 10.3390/cancers11122028. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

