Gene-Specific Control of tRNA Expression by RNA Polymerase II
- PMID: 32298650
- PMCID: PMC7273519
- DOI: 10.1016/j.molcel.2020.03.023
Gene-Specific Control of tRNA Expression by RNA Polymerase II
Abstract
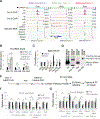
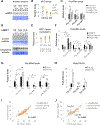
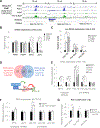
Increasing evidence suggests that tRNA levels are dynamically and specifically regulated in response to internal and external cues to modulate the cellular translational program. However, the molecular players and the mechanisms regulating the gene-specific expression of tRNAs are still unknown. Using an inducible auxin-degron system to rapidly deplete RPB1 (the largest subunit of RNA Pol II) in living cells, we identified Pol II as a direct gene-specific regulator of tRNA transcription. Our data suggest that Pol II transcription robustly interferes with Pol III function at specific tRNA genes. This activity was further found to be essential for MAF1-mediated repression of a large set of tRNA genes during serum starvation, indicating that repression of tRNA genes by Pol II is dynamically regulated. Hence, Pol II plays a direct and central role in the gene-specific regulation of tRNA expression.
Keywords: RNA Pol II; RNA Pol III; auxin degron; tRNA differential expression.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



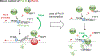
Similar articles
-
Maf1, a new player in the regulation of human RNA polymerase III transcription.PLoS One. 2006 Dec 27;1(1):e134. doi: 10.1371/journal.pone.0000134. PLoS One. 2006. PMID: 17205138 Free PMC article.
-
Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):320-329. doi: 10.1016/j.bbagrm.2018.01.011. Epub 2018 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29378333 Review.
-
RNA polymerase III under control: repression and de-repression.Trends Biochem Sci. 2011 Sep;36(9):451-6. doi: 10.1016/j.tibs.2011.06.008. Epub 2011 Aug 2. Trends Biochem Sci. 2011. PMID: 21816617
-
Casein kinase II-mediated phosphorylation of general repressor Maf1 triggers RNA polymerase III activation.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4926-31. doi: 10.1073/pnas.1010010108. Epub 2011 Mar 7. Proc Natl Acad Sci U S A. 2011. PMID: 21383183 Free PMC article.
-
Signaling to and from the RNA Polymerase III Transcription and Processing Machinery.Annu Rev Biochem. 2018 Jun 20;87:75-100. doi: 10.1146/annurev-biochem-062917-012624. Epub 2018 Jan 12. Annu Rev Biochem. 2018. PMID: 29328783 Free PMC article. Review.
Cited by
-
Non-Coding RNAs: Regulators of Stress, Ageing, and Developmental Decisions in Yeast?Cells. 2024 Mar 29;13(7):599. doi: 10.3390/cells13070599. Cells. 2024. PMID: 38607038 Free PMC article. Review.
-
Manipulation of RNA polymerase III by Herpes Simplex Virus-1.Nat Commun. 2022 Feb 2;13(1):623. doi: 10.1038/s41467-022-28144-8. Nat Commun. 2022. PMID: 35110532 Free PMC article.
-
Human Cytomegalovirus Infection Elicits Global Changes in Host Transcription by RNA Polymerases I, II, and III.Viruses. 2022 Apr 9;14(4):779. doi: 10.3390/v14040779. Viruses. 2022. PMID: 35458509 Free PMC article.
-
Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III.Genome Biol. 2020 Jul 2;21(1):158. doi: 10.1186/s13059-020-02067-3. Genome Biol. 2020. PMID: 32616013 Free PMC article.
-
The tRNA regulome in neurodevelopmental and neuropsychiatric disease.Mol Psychiatry. 2022 Aug;27(8):3204-3213. doi: 10.1038/s41380-022-01585-9. Epub 2022 May 3. Mol Psychiatry. 2022. PMID: 35505091 Free PMC article. Review.
References
-
- Arimbasseri GA (2018). Interactions between RNAP III transcription machinery and tRNA processing factors. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 1861, 354–360. - PubMed
-
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

