Charting the native architecture of Chlamydomonas thylakoid membranes with single-molecule precision
- PMID: 32297859
- PMCID: PMC7164959
- DOI: 10.7554/eLife.53740
Charting the native architecture of Chlamydomonas thylakoid membranes with single-molecule precision
Abstract
Thylakoid membranes scaffold an assortment of large protein complexes that work together to harness the energy of light. It has been a longstanding challenge to visualize how the intricate thylakoid network organizes these protein complexes to finely tune the photosynthetic reactions. Previously, we used in situ cryo-electron tomography to reveal the native architecture of thylakoid membranes (Engel et al., 2015). Here, we leverage technical advances to resolve the individual protein complexes within these membranes. Combined with a new method to visualize membrane surface topology, we map the molecular landscapes of thylakoid membranes inside green algae cells. Our tomograms provide insights into the molecular forces that drive thylakoid stacking and reveal that photosystems I and II are strictly segregated at the borders between appressed and non-appressed membrane domains. This new approach to charting thylakoid topology lays the foundation for dissecting photosynthetic regulation at the level of single protein complexes within the cell.
Keywords: chlamydomonas reinhardtii; chloroplast; electron microscopy; in situ; membranogram; molecular biophysics; photosynthesis; plant biology; structural biology; thylakoid.
© 2020, Wietrzynski et al.
Conflict of interest statement
WW, MS, DT, SA, AK, JP, WB, BE No competing interests declared
Figures

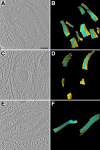
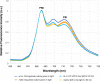




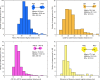



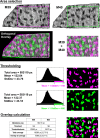

Similar articles
-
Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography.Elife. 2015 Jan 13;4:e04889. doi: 10.7554/eLife.04889. Elife. 2015. PMID: 25584625 Free PMC article.
-
Subdiffraction-resolution live-cell imaging for visualizing thylakoid membranes.Plant J. 2018 Oct;96(1):233-243. doi: 10.1111/tpj.14021. Epub 2018 Jul 30. Plant J. 2018. PMID: 29982996 Free PMC article.
-
Electron tomography of plant thylakoid membranes.J Exp Bot. 2011 Apr;62(7):2393-402. doi: 10.1093/jxb/err034. Epub 2011 Mar 25. J Exp Bot. 2011. PMID: 21441405 Review.
-
Electrokinetic and light scattering properties of pea and Chlamydomonas reinhardtii thylakoid membranes: effect of phytohemagglutinin.Electrophoresis. 2002 Jul;23(13):2138-43. doi: 10.1002/1522-2683(200207)23:13<2138::AID-ELPS2138>3.0.CO;2-P. Electrophoresis. 2002. PMID: 12210269
-
A brief history of how microscopic studies led to the elucidation of the 3D architecture and macromolecular organization of higher plant thylakoids.Photosynth Res. 2020 Sep;145(3):237-258. doi: 10.1007/s11120-020-00782-3. Epub 2020 Oct 5. Photosynth Res. 2020. PMID: 33017036 Free PMC article. Review.
Cited by
-
The Main Structural and Functional Characteristics of Photosystem-II-Enriched Membranes Isolated from Wild Type and cia3 Mutant Chlamydomonas reinhardtii.Life (Basel). 2020 May 14;10(5):63. doi: 10.3390/life10050063. Life (Basel). 2020. PMID: 32423065 Free PMC article.
-
In situ cryo-ET structure of phycobilisome-photosystem II supercomplex from red alga.Elife. 2021 Sep 13;10:e69635. doi: 10.7554/eLife.69635. Elife. 2021. PMID: 34515634 Free PMC article.
-
Morphological bases of phytoplankton energy management and physiological responses unveiled by 3D subcellular imaging.Nat Commun. 2021 Feb 16;12(1):1049. doi: 10.1038/s41467-021-21314-0. Nat Commun. 2021. PMID: 33594064 Free PMC article.
-
High light-induced changes in whole-cell proteomic profile and its correlation with the organization of thylakoid super-complex in cyclic electron transport mutants of Chlamydomonas reinhardtii.Front Plant Sci. 2023 Jul 7;14:1198474. doi: 10.3389/fpls.2023.1198474. eCollection 2023. Front Plant Sci. 2023. PMID: 37521924 Free PMC article.
-
Cryogenic Super-Resolution Fluorescence and Electron Microscopy Correlated at the Nanoscale.Annu Rev Phys Chem. 2021 Apr 20;72:253-278. doi: 10.1146/annurev-physchem-090319-051546. Epub 2021 Jan 13. Annu Rev Phys Chem. 2021. PMID: 33441030 Free PMC article. Review.
References
-
- Albanese P, Melero R, Engel BD, Grinzato A, Berto P, Manfredi M, Chiodoni A, Vargas J, Sorzano CÓS, Marengo E, Saracco G, Zanotti G, Carazo JM, Pagliano C. Pea PSII-LHCII supercomplexes form pairs by making connections across the stromal gap. Scientific Reports. 2017;7:10067. doi: 10.1038/s41598-017-10700-8. - DOI - PMC - PubMed
-
- Anderson JM. The grana margins of plant thylakoid membranes. Physiologia Plantarum. 1989;76:243–248. doi: 10.1111/j.1399-3054.1989.tb05640.x. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

