hnRNPA2B1 Associated with Recruitment of RNA into Exosomes Plays a Key Role in Herpes Simplex Virus 1 Release from Infected Cells
- PMID: 32295924
- PMCID: PMC7307161
- DOI: 10.1128/JVI.00367-20
hnRNPA2B1 Associated with Recruitment of RNA into Exosomes Plays a Key Role in Herpes Simplex Virus 1 Release from Infected Cells
Abstract
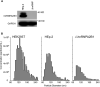
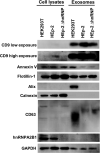
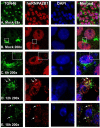
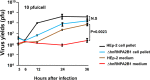
hnRNPA2B1, an abundant cellular protein, has been reported to recruit RNAs bearing a specific sequence (EXO motif) into exosomes. We characterized an exosome population averaging 100 ± 50 nm in diameter and containing a defined set of constitutive exosome markers. This population packages microRNAs (miRNAs) and can be directed to block targeted gene expression in a dose-dependent fashion. The objective of this study was to characterize the role of hnRNPA2B1 in the recruitment of miRNA. We report the following four key findings. (i) hnRNPA2B1 is not a component of exosomes produced in HEp-2 or HEK293T cells. Hence, hnRNPA2B1 carries its cargo, at most, to the site of exosome assembly, but it is not itself incorporated into exosomes. (ii) The accumulation of exosomes produced by cells in which the gene encoding hnRNPA2B1 has been knocked out (ΔhnRNPA2B1 cells) was reduced 3-fold. (iii) In uninfected HEp-2 cells, hnRNPA2B1 is localized in the nucleus. In cells infected with herpes simplex virus 1 (HSV-1), hnRNPA2B1 was quantitatively exported to the cytoplasm and at least a fraction of hnRNPA2B1 colocalized with a Golgi marker. (iv) Lastly, in ΔhnRNPA2B1 cells, there was a 2- to 3-fold reduction in virus yield but a significant (>10-fold) reduction in HSV-1 released through the apical surface into the extracellular environment. The absence of hnRNPA2B1 had no significant impact on the basolateral export of HSV-1 from infected to uninfected cells by direct cell-to-cell contact. The results suggest that hnRNPA2B1 plays a key role in the transport of enveloped virus from its site of assembly to the extracellular environment.IMPORTANCE In this report, we show that hnRNPA2B1 is not a component of exosomes produced in HEp-2 or HEK293T cells. In herpes simplex virus 1 (HSV-1)-infected cells, hnRNPA2B1 was quantitatively translocated from the nucleus into the cytoplasm. In infected ΔhnRNPA2B1 cells, Golgi-dependent transport of virus from the apical surface to the extracellular medium was significantly reduced. In essence, this report supports the hypothesis that hnRNPA2B1 plays a key role in the egress of exosomes and HSV-1 from infected cells.
Keywords: HSV-1; exosome; hnRNPA2B1; release.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):E4991-6. doi: 10.1073/pnas.1419338111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368198 Free PMC article.
-
Herpes Simplex Virus 1 MicroRNA miR-H28 Exported to Uninfected Cells in Exosomes Restricts Cell-to-Cell Virus Spread by Inducing Gamma Interferon mRNA.J Virol. 2019 Oct 15;93(21):e01005-19. doi: 10.1128/JVI.01005-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31413129 Free PMC article.
-
miRNAs Targeting ICP4 and Delivered to Susceptible Cells in Exosomes Block HSV-1 Replication in a Dose-Dependent Manner.Mol Ther. 2018 Apr 4;26(4):1032-1039. doi: 10.1016/j.ymthe.2018.02.016. Epub 2018 Feb 21. Mol Ther. 2018. PMID: 29526650 Free PMC article.
-
Herpesviruses hijack host exosomes for viral pathogenesis.Semin Cell Dev Biol. 2017 Jul;67:91-100. doi: 10.1016/j.semcdb.2017.03.005. Epub 2017 Apr 26. Semin Cell Dev Biol. 2017. PMID: 28456604 Review.
-
HSV-1 Cytoplasmic Envelopment and Egress.Int J Mol Sci. 2020 Aug 19;21(17):5969. doi: 10.3390/ijms21175969. Int J Mol Sci. 2020. PMID: 32825127 Free PMC article. Review.
Cited by
-
Culture conditions greatly impact the levels of vesicular and extravesicular Ago2 and RNA in extracellular vesicle preparations.J Extracell Vesicles. 2023 Nov;12(11):e12366. doi: 10.1002/jev2.12366. J Extracell Vesicles. 2023. PMID: 37885043 Free PMC article.
-
Release of HSV-1 Cell-Free Virions: Mechanisms, Regulation, and Likely Role in Human-Human Transmission.Viruses. 2021 Nov 30;13(12):2395. doi: 10.3390/v13122395. Viruses. 2021. PMID: 34960664 Free PMC article. Review.
-
Molecular mechanisms underlying host-induced gene silencing.Plant Cell. 2022 Aug 25;34(9):3183-3199. doi: 10.1093/plcell/koac165. Plant Cell. 2022. PMID: 35666177 Free PMC article. Review.
-
Mechanism of action of hepatitis B virus S antigen transport-inhibiting oligonucleotide polymer, STOPS, molecules.Mol Ther Nucleic Acids. 2021 Dec 11;27:335-348. doi: 10.1016/j.omtn.2021.12.013. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2021. PMID: 35024245 Free PMC article.
-
Exosomal MicroRNAs Mediating Crosstalk Between Cancer Cells With Cancer-Associated Fibroblasts and Tumor-Associated Macrophages in the Tumor Microenvironment.Front Oncol. 2021 Apr 1;11:631703. doi: 10.3389/fonc.2021.631703. eCollection 2021. Front Oncol. 2021. PMID: 33869017 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

