M2-like, dermal macrophages are maintained via IL-4/CCL24-mediated cooperative interaction with eosinophils in cutaneous leishmaniasis
- PMID: 32276966
- PMCID: PMC7385908
- DOI: 10.1126/sciimmunol.aaz4415
M2-like, dermal macrophages are maintained via IL-4/CCL24-mediated cooperative interaction with eosinophils in cutaneous leishmaniasis
Abstract
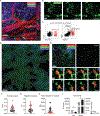
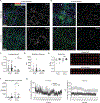
Tissue-resident macrophages (TRMs) maintain tissue homeostasis, but they can also provide a replicative niche for intracellular pathogens such as Leishmania How dermal TRMs proliferate and maintain their M2 properties even in the strong TH1 environment of the L. major infected dermis is not clear. Here, we show that, in infected mice lacking IL-4/13 from eosinophils, dermal TRMs shifted to a proinflammatory state, their numbers declined, and disease was attenuated. Intravital microscopy revealed a rapid infiltration of eosinophils followed by their tight interaction with dermal TRMs. IL-4-stimulated dermal TRMs, in concert with IL-10, produced a large amount of CCL24, which functioned to amplify eosinophil influx and their interaction with dermal TRMs. An intraperitoneal helminth infection model also demonstrated a requirement for eosinophil-derived IL-4 to maintain tissue macrophages through a CCL24-mediated amplification loop. CCL24 secretion was confined to resident macrophages in other tissues, implicating eosinophil-TRM cooperative interactions in diverse inflammatory settings.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures





Similar articles
-
Dermis resident macrophages orchestrate localized ILC2-eosinophil circuitries to maintain their M2-like properties and promote non-healing cutaneous leishmaniasis.Res Sq [Preprint]. 2023 Apr 5:rs.3.rs-2644705. doi: 10.21203/rs.3.rs-2644705/v1. Res Sq. 2023. Update in: Nat Commun. 2023 Nov 29;14(1):7852. doi: 10.1038/s41467-023-43588-2 PMID: 37066418 Free PMC article. Updated. Preprint.
-
Dermis resident macrophages orchestrate localized ILC2 eosinophil circuitries to promote non-healing cutaneous leishmaniasis.Nat Commun. 2023 Nov 29;14(1):7852. doi: 10.1038/s41467-023-43588-2. Nat Commun. 2023. PMID: 38030609 Free PMC article.
-
IL-10 enhances the phenotype of M2 macrophages induced by IL-4 and confers the ability to increase eosinophil migration.Int Immunol. 2015 Mar;27(3):131-41. doi: 10.1093/intimm/dxu090. Epub 2014 Sep 29. Int Immunol. 2015. PMID: 25267883
-
Immune serum-activated human macrophages coordinate with eosinophils to immobilize Ascaris suum larvae.Parasite Immunol. 2020 Jul;42(7):e12728. doi: 10.1111/pim.12728. Epub 2020 May 25. Parasite Immunol. 2020. PMID: 32394439 Review.
-
Non-canonical alternatives: what a macrophage is 4.J Exp Med. 2012 Mar 12;209(3):427-31. doi: 10.1084/jem.20120295. J Exp Med. 2012. PMID: 22412174 Free PMC article. Review.
Cited by
-
Recent advances in understanding the role of eosinophils.Fac Rev. 2022 Sep 27;11:26. doi: 10.12703/r/11-26. eCollection 2022. Fac Rev. 2022. PMID: 36225210 Free PMC article. Review.
-
Eosinophils, but Not Type 2 Innate Lymphoid Cells, Are the Predominant Source of Interleukin 4 during the Innate Phase of Leishmania major Infection.Pathogens. 2022 Jul 25;11(8):828. doi: 10.3390/pathogens11080828. Pathogens. 2022. PMID: 35894051 Free PMC article.
-
Essential Role of Enzymatic Activity in the Leishmanicidal Mechanism of the Eosinophil Cationic Protein (RNase 3).ACS Infect Dis. 2022 Jul 8;8(7):1207-1217. doi: 10.1021/acsinfecdis.1c00537. Epub 2022 Jun 22. ACS Infect Dis. 2022. PMID: 35731709 Free PMC article.
-
Hepatic macrophages play critical roles in the establishment and growth of hydatid cysts in the liver during Echinococcus granulosus sensu stricto infection.PLoS Negl Trop Dis. 2023 Nov 6;17(11):e0011746. doi: 10.1371/journal.pntd.0011746. eCollection 2023 Nov. PLoS Negl Trop Dis. 2023. PMID: 37930989 Free PMC article.
-
High-speed, three-dimensional imaging reveals chemotactic behaviour specific to human-infective Leishmania parasites.Elife. 2021 Jun 28;10:e65051. doi: 10.7554/eLife.65051. Elife. 2021. PMID: 34180835 Free PMC article.
References
-
- Ginhoux F, Guilliams M, Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44, 439–449 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

