Bioenergetics of the Dictyostelium Kinesin-8 Motor Isoform
- PMID: 32272590
- PMCID: PMC7226124
- DOI: 10.3390/biom10040563
Bioenergetics of the Dictyostelium Kinesin-8 Motor Isoform
Abstract
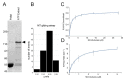
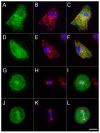
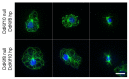
The functional organization of microtubules in eukaryotic cells requires a combination of their inherent dynamic properties, interactions with motor machineries, and interactions with accessory proteins to affect growth, shrinkage, stability, and architecture. In most organisms, the Kinesin-8 family of motors play an integral role in these organizations, well known for their mitotic activities in microtubule (MT) length control and kinetochore interactions. In Dictyostelium discoideum, the function of Kinesin-8 remains elusive. We present here some biochemical properties and localization data that indicate that this motor (DdKif10) shares some motility properties with other Kinesin-8s but also illustrates differences in microtubule localization and depolymerase action that highlight functional diversity.
Keywords: Dictyostelium; cell motility; kinesin; microtubules.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Microtubule-nucleus interactions in Dictyostelium discoideum mediated by central motor kinesins.Eukaryot Cell. 2009 May;8(5):723-31. doi: 10.1128/EC.00018-09. Epub 2009 Mar 13. Eukaryot Cell. 2009. PMID: 19286984 Free PMC article.
-
A non-mitotic CENP-E homolog in Dictyostelium discoideum with slow motor activity.Biochem Biophys Res Commun. 2013 Feb 15;431(3):490-5. doi: 10.1016/j.bbrc.2013.01.030. Epub 2013 Jan 16. Biochem Biophys Res Commun. 2013. PMID: 23333327
-
Properties of the Kinesin-1 motor DdKif3 from Dictyostelium discoideum.Eur J Cell Biol. 2008 Apr;87(4):237-49. doi: 10.1016/j.ejcb.2007.11.001. Epub 2007 Dec 21. Eur J Cell Biol. 2008. PMID: 18160177
-
13 Plus 1: A 30-Year Perspective on Microtubule-Based Motility in Dictyostelium.Cells. 2020 Feb 25;9(3):528. doi: 10.3390/cells9030528. Cells. 2020. PMID: 32106406 Free PMC article. Review.
-
Kinesin motors and microtubule-based organelle transport in Dictyostelium discoideum.J Muscle Res Cell Motil. 2002;23(7-8):631-8. doi: 10.1023/a:1024403006680. J Muscle Res Cell Motil. 2002. PMID: 12952062 Review.
Cited by
-
These motors were made for walking.Protein Sci. 2020 Aug;29(8):1707-1723. doi: 10.1002/pro.3895. Epub 2020 Jun 26. Protein Sci. 2020. PMID: 32472639 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

