Genome-wide analysis of short interspersed nuclear elements provides insight into gene and genome evolution in citrus
- PMID: 32271875
- PMCID: PMC7315354
- DOI: 10.1093/dnares/dsaa004
Genome-wide analysis of short interspersed nuclear elements provides insight into gene and genome evolution in citrus
Abstract
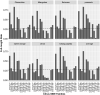
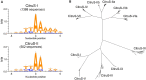
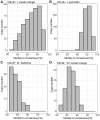
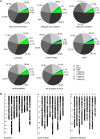
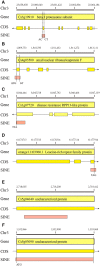
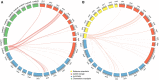
Short interspersed nuclear elements (SINEs) are non-autonomous retrotransposons that are highly abundant, but not well annotated, in plant genomes. In this study, we identified 41,573 copies of SINEs in seven citrus genomes, including 11,275 full-length copies. The citrus SINEs were distributed among 12 families, with an average full-length rate of 0.27, and were dispersed throughout the chromosomes, preferentially in AT-rich areas. Approximately 18.4% of citrus SINEs were found in close proximity (≤1 kb upstream) to genes, indicating a significant enrichment of SINEs in promoter regions. Citrus SINEs promote gene and genome evolution by offering exons as well as splice sites and start and stop codons, creating novel genes and forming tandem and dispersed repeat structures. Comparative analysis of unique homologous SINE-containing loci (HSCLs) revealed chromosome rearrangements in sweet orange, pummelo, and mandarin, suggesting that unique HSCLs might be valuable for understanding chromosomal abnormalities. This study of SINEs provides us with new perspectives and new avenues by which to understand the evolution of citrus genes and genomes.
Keywords: citrus; evolution; gene association; genome; short interspersed nuclear elements.
© The Author(s) 2020. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures

Similar articles
-
Diversification, evolution and methylation of short interspersed nuclear element families in sugar beet and related Amaranthaceae species.Plant J. 2016 Jan;85(2):229-44. doi: 10.1111/tpj.13103. Plant J. 2016. PMID: 26676716
-
Short interspersed nuclear elements (SINEs) are abundant in Solanaceae and have a family-specific impact on gene structure and genome organization.Plant J. 2016 May;86(3):268-85. doi: 10.1111/tpj.13170. Plant J. 2016. PMID: 26996788
-
Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes.Plant Cell. 2011 Sep;23(9):3117-28. doi: 10.1105/tpc.111.088682. Epub 2011 Sep 9. Plant Cell. 2011. PMID: 21908723 Free PMC article.
-
SINEs.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):772-86. doi: 10.1002/wrna.91. Epub 2011 Jul 7. Wiley Interdiscip Rev RNA. 2011. PMID: 21976282 Review.
-
SINEs as driving forces in genome evolution.Genome Dyn. 2012;7:92-107. doi: 10.1159/000337117. Epub 2012 Jun 25. Genome Dyn. 2012. PMID: 22759815 Review.
Cited by
-
Genetic Evaluation and Population Structure of Jiangsu Native Pigs in China Revealed by SINE Insertion Polymorphisms.Animals (Basel). 2022 May 25;12(11):1345. doi: 10.3390/ani12111345. Animals (Basel). 2022. PMID: 35681812 Free PMC article.
-
Accelerating de novo SINE annotation in plant and animal genomes.Mob DNA. 2024 Oct 19;15(1):24. doi: 10.1186/s13100-024-00331-y. Mob DNA. 2024. PMID: 39427206 Free PMC article.
References
-
- Naito K., Zhang F., Tsukiyama T., et al.2009, Unexpected consequences of a sudden and massive transposon amplification on rice gene expression, Nature, 461, 1130–4. - PubMed
-
- Wang L., He F., Huang Y., et al.2018, Genome of wild mandarin and domestication history of mandarin, Mol. Plant, 11, 1024–37. - PubMed
-
- Zhang J., Yu C., Krishnaswamy L., Peterson T. 2011, Transposable elements as catalysts for chromosome rearrangements. In: Birchler J.A., ed., Plant Chromosome Engineering: Methods and Protocols, Humana Press: Totowa, NJ, pp. 315–26. - PubMed
-
- Feinberg A.P. 2007, Phenotypic plasticity and the epigenetics of human disease, Nature, 447, 433–40. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

