Hepatitis C virus infection and tight junction proteins: The ties that bind
- PMID: 32268133
- PMCID: PMC7613427
- DOI: 10.1016/j.bbamem.2020.183296
Hepatitis C virus infection and tight junction proteins: The ties that bind
Abstract
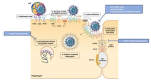
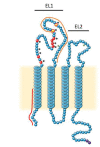
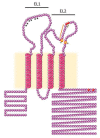
The hepatitis C virus (HCV) is a major cause of liver diseases ranging from liver inflammation to advanced liver diseases like cirrhosis and hepatocellular carcinoma (HCC). HCV infection is restricted to the liver, and more specifically to hepatocytes, which represent around 80% of liver cells. The mechanism of HCV entry in human hepatocytes has been extensively investigated since the discovery of the virus 30 years ago. The entry mechanism is a multi-step process relying on several host factors including heparan sulfate proteoglycan (HSPG), low density lipoprotein receptor (LDLR), tetraspanin CD81, Scavenger Receptor class B type I (SR-BI), Epidermal Growth Factor Receptor (EGFR) and Niemann-Pick C1-like 1 (NPC1L1). Moreover, in order to establish a persistent infection, HCV entry is dependent on the presence of tight junction (TJ) proteins Claudin-1 (CLDN1) and Occludin (OCLN). In the liver, tight junction proteins play a role in architecture and homeostasis including sealing the apical pole of adjacent cells to form bile canaliculi and separating the basolateral domain drained by sinusoidal blood flow. In this review, we will highlight the role of liver tight junction proteins in HCV infection, and we will discuss the potential targeted therapeutic approaches to improve virus eradication.
Keywords: Claudin-1; Hepatitis C virus; Occludin; Tight junctions; Viral entry and spreading.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Inserm, the University of Strasbourg and the Strasbourg University Hospitals have filed patent applications for the use of anti-Claudin1 antibodies for treatment of HCV infection which have been licensed to Alentis Therapeutics, Basel, Switzerland.
Figures
Similar articles
-
Single Particle Imaging of Polarized Hepatoma Organoids upon Hepatitis C Virus Infection Reveals an Ordered and Sequential Entry Process.Cell Host Microbe. 2018 Mar 14;23(3):382-394.e5. doi: 10.1016/j.chom.2018.02.005. Cell Host Microbe. 2018. PMID: 29544098 Free PMC article.
-
Infection with hepatitis C virus depends on TACSTD2, a regulator of claudin-1 and occludin highly downregulated in hepatocellular carcinoma.PLoS Pathog. 2018 Mar 14;14(3):e1006916. doi: 10.1371/journal.ppat.1006916. eCollection 2018 Mar. PLoS Pathog. 2018. PMID: 29538454 Free PMC article.
-
Mice Expressing Minimally Humanized CD81 and Occludin Genes Support Hepatitis C Virus Uptake In Vivo.J Virol. 2017 Jan 31;91(4):e01799-16. doi: 10.1128/JVI.01799-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928007 Free PMC article.
-
How hepatitis C virus invades hepatocytes: the mystery of viral entry.World J Gastroenterol. 2014 Apr 7;20(13):3457-67. doi: 10.3748/wjg.v20.i13.3457. World J Gastroenterol. 2014. PMID: 24707128 Free PMC article. Review.
-
The hepatitis C virus and its hepatic environment: a toxic but finely tuned partnership.Biochem J. 2009 Oct 12;423(3):303-14. doi: 10.1042/BJ20091000. Biochem J. 2009. PMID: 19807698 Review.
Cited by
-
Occludin: a gatekeeper of brain Infection by HIV-1.Fluids Barriers CNS. 2023 Oct 16;20(1):73. doi: 10.1186/s12987-023-00476-7. Fluids Barriers CNS. 2023. PMID: 37840143 Free PMC article. Review.
-
Micronutrients at Supplemental Levels, Tight Junctions and Epithelial Barrier Function: A Narrative Review.Int J Mol Sci. 2024 Mar 19;25(6):3452. doi: 10.3390/ijms25063452. Int J Mol Sci. 2024. PMID: 38542424 Free PMC article. Review.
-
The Basic Requirement of Tight Junction Proteins in Blood-Brain Barrier Function and Their Role in Pathologies.Int J Mol Sci. 2024 May 21;25(11):5601. doi: 10.3390/ijms25115601. Int J Mol Sci. 2024. PMID: 38891789 Free PMC article. Review.
-
Contemporary Insights into Hepatitis C Virus: A Comprehensive Review.Microorganisms. 2024 May 21;12(6):1035. doi: 10.3390/microorganisms12061035. Microorganisms. 2024. PMID: 38930417 Free PMC article. Review.
-
TM4SF5-Mediated Regulation of Hepatocyte Transporters during Metabolic Liver Diseases.Int J Mol Sci. 2022 Jul 29;23(15):8387. doi: 10.3390/ijms23158387. Int J Mol Sci. 2022. PMID: 35955521 Free PMC article. Review.
References
-
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. - PubMed
-
- Lindenbach BD, Thiel HJ, Rice CM. In: Flaviviridae: The viruses and their replication, Fields Virology. 5th Edition. Knipe DM, Howley PM, editors. Vol. 2007. Lippincott-Raven Publishers; Philadelphia: 2007. pp. 1101–1152.
-
- Poordad F, Dieterich D. Treating hepatitis C: current standard of care and emerging direct-acting antiviral agents. J Viral Hepat. 2012;19:449–464. - PubMed
-
- Das D, Pandya M. Recent Advancement of Direct-acting Antiviral Agents (DAAs) in Hepatitis C Therapy. Mini Rev Med Chem. 2018;18:584–596. - PubMed
-
- Loo N, Hanysak B, Mann J, Ramirez R, Kim J, Mitchell R, Van Frank T, Guerrero R, Hinojosa K, Christensen K, Pedicone LD, et al. Real-world observational experience with direct-acting antivirals for hepatitis C: baseline resistance, efficacy, and need for long-term surveillance. Medicine (Baltimore) 2019;98:e16254. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

