The splicing-factor Prp40 affects dynein-dynactin function in Aspergillus nidulans
- PMID: 32267207
- PMCID: PMC7353152
- DOI: 10.1091/mbc.E20-03-0166
The splicing-factor Prp40 affects dynein-dynactin function in Aspergillus nidulans
Abstract
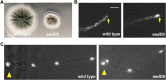
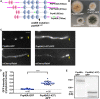
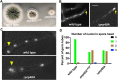
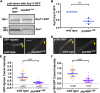
The multi-component cytoplasmic dynein transports cellular cargoes with the help of another multi-component complex dynactin, but we do not know enough about factors that may affect the assembly and functions of these proteins. From a genetic screen for mutations affecting early-endosome distribution in Aspergillus nidulans, we identified the prp40AL438* mutation in Prp40A, a homologue of Prp40, an essential RNA-splicing factor in the budding yeast. Prp40A is not essential for splicing, although it associates with the nuclear splicing machinery. The prp40AL438* mutant is much healthier than the ∆prp40A mutant, but both mutants exhibit similar defects in dynein-mediated early-endosome transport and nuclear distribution. In the prp40AL438* mutant, the frequency but not the speed of dynein-mediated early-endosome transport is decreased, which correlates with a decrease in the microtubule plus-end accumulations of dynein and dynactin. Within the dynactin complex, the actin-related protein Arp1 forms a mini-filament. In a pull-down assay, the amount of Arp1 pulled down with its pointed-end protein Arp11 is lowered in the prp40AL438* mutant. In addition, we found from published interactome data that a mammalian Prp40 homologue PRPF40A interacts with Arp1. Thus, Prp40 homologues may regulate the assembly or function of dynein-dynactin and their mechanisms deserve to be further studied.
Figures
Similar articles
-
The actin capping protein in Aspergillus nidulans enhances dynein function without significantly affecting Arp1 filament assembly.Sci Rep. 2018 Jul 30;8(1):11419. doi: 10.1038/s41598-018-29818-4. Sci Rep. 2018. PMID: 30061726 Free PMC article.
-
p25 of the dynactin complex plays a dual role in cargo binding and dynactin regulation.J Biol Chem. 2018 Oct 5;293(40):15606-15619. doi: 10.1074/jbc.RA118.004000. Epub 2018 Aug 24. J Biol Chem. 2018. PMID: 30143531 Free PMC article.
-
Arp11 affects dynein-dynactin interaction and is essential for dynein function in Aspergillus nidulans.Traffic. 2008 Jul;9(7):1073-87. doi: 10.1111/j.1600-0854.2008.00748.x. Epub 2008 Apr 11. Traffic. 2008. PMID: 18410488 Free PMC article.
-
The role of the dynactin complex in intracellular motility.Int Rev Cytol. 1998;182:69-109. doi: 10.1016/s0074-7696(08)62168-3. Int Rev Cytol. 1998. PMID: 9522459 Review.
-
Cytoplasmic dynein and dynactin in cell division and intracellular transport.Curr Opin Cell Biol. 1999 Feb;11(1):45-53. doi: 10.1016/s0955-0674(99)80006-4. Curr Opin Cell Biol. 1999. PMID: 10047518 Review.
Cited by
-
VezA/vezatin facilitates proper assembly of the dynactin complex in vivo.bioRxiv [Preprint]. 2024 Apr 20:2024.04.19.590248. doi: 10.1101/2024.04.19.590248. bioRxiv. 2024. Update in: Cell Rep. 2024 Nov 26;43(11):114943. doi: 10.1016/j.celrep.2024.114943 PMID: 38659795 Free PMC article. Updated. Preprint.
-
Kinesin-1 autoinhibition facilitates the initiation of dynein cargo transport.J Cell Biol. 2023 Mar 6;222(3):e202205136. doi: 10.1083/jcb.202205136. Epub 2022 Dec 16. J Cell Biol. 2023. PMID: 36524956 Free PMC article.
-
Cargo-Mediated Activation of Cytoplasmic Dynein in vivo.Front Cell Dev Biol. 2020 Oct 23;8:598952. doi: 10.3389/fcell.2020.598952. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195284 Free PMC article. Review.
References
-
- Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Peñalva MA. (2009). Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic , 57–75. - PubMed
-
- Abu Dayyeh BK, Quan TK, Castro M, Ruby SW. (2002). Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J Biol Chem , 20221–20233. - PubMed
-
- Allen M, Friedler A, Schon O, Bycroft M. (2002). The structure of an FF domain from human HYPA/FBP11. J Mol Biol , 411–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

