Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus
- PMID: 32265329
- PMCID: PMC7157774
- DOI: 10.1128/mBio.00524-20
Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus
Abstract
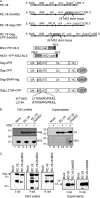
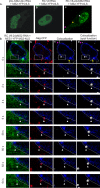
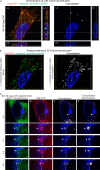
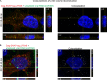
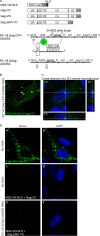
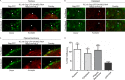
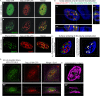
Packaging of genomic RNA (gRNA) by retroviruses is essential for infectivity, yet the subcellular site of the initial interaction between the Gag polyprotein and gRNA remains poorly defined. Because retroviral particles are released from the plasma membrane, it was previously thought that Gag proteins initially bound to gRNA in the cytoplasm or at the plasma membrane. However, the Gag protein of the avian retrovirus Rous sarcoma virus (RSV) undergoes active nuclear trafficking, which is required for efficient gRNA encapsidation (L. Z. Scheifele, R. A. Garbitt, J. D. Rhoads, and L. J. Parent, Proc Natl Acad Sci U S A 99:3944-3949, 2002, https://doi.org/10.1073/pnas.062652199; R. Garbitt-Hirst, S. P. Kenney, and L. J. Parent, J Virol 83:6790-6797, 2009, https://doi.org/10.1128/JVI.00101-09). These results raise the intriguing possibility that the primary contact between Gag and gRNA might occur in the nucleus. To examine this possibility, we created a RSV proviral construct that includes 24 tandem repeats of MS2 RNA stem-loops, making it possible to track RSV viral RNA (vRNA) in live cells in which a fluorophore-conjugated MS2 coat protein is coexpressed. Using confocal microscopy, we observed that both wild-type Gag and a nuclear export mutant (Gag.L219A) colocalized with vRNA in the nucleus. In live-cell time-lapse images, the wild-type Gag protein trafficked together with vRNA as a single ribonucleoprotein (RNP) complex in the nucleoplasm near the nuclear periphery, appearing to traverse the nuclear envelope into the cytoplasm. Furthermore, biophysical imaging methods suggest that Gag and the unspliced vRNA physically interact in the nucleus. Taken together, these data suggest that RSV Gag binds unspliced vRNA to export it from the nucleus, possibly for packaging into virions as the viral genome.IMPORTANCE Retroviruses cause severe diseases in animals and humans, including cancer and acquired immunodeficiency syndromes. To propagate infection, retroviruses assemble new virus particles that contain viral proteins and unspliced vRNA to use as gRNA. Despite the critical requirement for gRNA packaging, the molecular mechanisms governing the identification and selection of gRNA by the Gag protein remain poorly understood. In this report, we demonstrate that the Rous sarcoma virus (RSV) Gag protein colocalizes with unspliced vRNA in the nucleus in the interchromatin space. Using live-cell confocal imaging, RSV Gag and unspliced vRNA were observed to move together from inside the nucleus across the nuclear envelope, suggesting that the Gag-gRNA complex initially forms in the nucleus and undergoes nuclear export into the cytoplasm as a viral ribonucleoprotein (vRNP) complex.
Keywords: Gag proteins; RNA trafficking; Rous sarcoma virus; genomic RNA packaging; live cell imaging; nucleocytoplasmic trafficking; retrovirus assembly.
Copyright © 2020 Maldonado et al.
Figures
Similar articles
-
An Infectious Rous Sarcoma Virus Gag Mutant That Is Defective in Nuclear Cycling.J Virol. 2021 Sep 27;95(20):e0064821. doi: 10.1128/JVI.00648-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319154 Free PMC article.
-
Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA.Proc Natl Acad Sci U S A. 2010 May 18;107(20):9358-63. doi: 10.1073/pnas.1000304107. Epub 2010 Apr 30. Proc Natl Acad Sci U S A. 2010. PMID: 20435918 Free PMC article.
-
Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging.J Virol. 2009 Jul;83(13):6790-7. doi: 10.1128/JVI.00101-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369339 Free PMC article.
-
Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors.Viruses. 2016 Sep 20;8(9):257. doi: 10.3390/v8090257. Viruses. 2016. PMID: 27657110 Free PMC article. Review.
-
Visualization of Retroviral Gag-Genomic RNA Cellular Interactions Leading to Genome Encapsidation and Viral Assembly: An Overview.Viruses. 2022 Feb 5;14(2):324. doi: 10.3390/v14020324. Viruses. 2022. PMID: 35215917 Free PMC article. Review.
Cited by
-
Retroviral RNA Processing.Viruses. 2022 May 23;14(5):1113. doi: 10.3390/v14051113. Viruses. 2022. PMID: 35632854 Free PMC article. Review.
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. Retrovirology. 2024. PMID: 38898526 Free PMC article.
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Epitranscriptomic regulation of HIV-1 full-length RNA packaging.Nucleic Acids Res. 2022 Feb 28;50(4):2302-2318. doi: 10.1093/nar/gkac062. Nucleic Acids Res. 2022. PMID: 35137199 Free PMC article.
-
Unveiling the DHX15-G-patch interplay in retroviral RNA packaging.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2407990121. doi: 10.1073/pnas.2407990121. Epub 2024 Sep 25. Proc Natl Acad Sci U S A. 2024. PMID: 39320912 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

