Major gene expression changes and epigenetic remodelling in Nile tilapia muscle after just one generation of domestication
- PMID: 32264748
- PMCID: PMC7116051
- DOI: 10.1080/15592294.2020.1748914
Major gene expression changes and epigenetic remodelling in Nile tilapia muscle after just one generation of domestication
Abstract
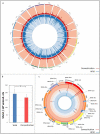
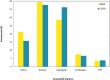
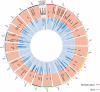
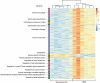
The historically recent domestication of fishes has been essential to meet the protein demands of a growing human population. Selection for traits of interest during domestication is a complex process whose epigenetic basis is poorly understood. Cytosine hydroxymethylation is increasingly recognized as an important DNA modification involved in epigenetic regulation. In the present study, we investigated if hydroxymethylation plays a role in fish domestication and demonstrated for the first time at a genome-wide level and single nucleotide resolution that the muscle hydroxymethylome changes after a single generation of Nile tilapia (Oreochromis niloticus, Linnaeus) domestication. The overall decrease in hydroxymethylcytosine levels was accompanied by the downregulation of 2015 genes in fish reared in captivity compared to their wild progenitors. In contrast, several myogenic and metabolic genes that can affect growth potential were upregulated. There were 126 differentially hydroxymethylated cytosines between groups, which were not due to genetic variation; they were associated with genes involved in immune-, growth- and neuronal-related pathways. Taken together, our data unveil a new role for DNA hydroxymethylation in epigenetic regulation of fish domestication with impact in aquaculture and implications in artificial selection, environmental adaptation and genome evolution.
Keywords: Oreochromis niloticus; DNA hydroxymethylation; Domestication; epigenetics; muscle growth.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
DNA hydroxymethylation differences underlie phenotypic divergence of somatic growth in Nile tilapia reared in common garden.Epigenetics. 2023 Dec;18(1):2282323. doi: 10.1080/15592294.2023.2282323. Epub 2023 Nov 27. Epigenetics. 2023. PMID: 38010265 Free PMC article.
-
Genome-wide hydroxymethylation profiles in liver of female Nile tilapia with distinct growth performance.Sci Data. 2023 Mar 1;10(1):114. doi: 10.1038/s41597-023-01996-5. Sci Data. 2023. PMID: 36859394 Free PMC article.
-
Epigenetic mapping of the somatotropic axis in Nile tilapia reveals differential DNA hydroxymethylation marks associated with growth.Genomics. 2021 Sep;113(5):2953-2964. doi: 10.1016/j.ygeno.2021.06.037. Epub 2021 Jun 30. Genomics. 2021. PMID: 34214627 Free PMC article.
-
Environmental Epigenetics and Genome Flexibility: Focus on 5-Hydroxymethylcytosine.Int J Mol Sci. 2020 May 2;21(9):3223. doi: 10.3390/ijms21093223. Int J Mol Sci. 2020. PMID: 32370155 Free PMC article. Review.
-
Hydroxymethylation and tumors: can 5-hydroxymethylation be used as a marker for tumor diagnosis and treatment?Hum Genomics. 2020 May 6;14(1):15. doi: 10.1186/s40246-020-00265-5. Hum Genomics. 2020. PMID: 32375881 Free PMC article. Review.
Cited by
-
The novel circular RNA CircMef2c is positively associated with muscle growth in Nile tilapia.Genomics. 2023 May;115(3):110598. doi: 10.1016/j.ygeno.2023.110598. Epub 2023 Mar 9. Genomics. 2023. PMID: 36906188 Free PMC article.
-
DNA hydroxymethylation differences underlie phenotypic divergence of somatic growth in Nile tilapia reared in common garden.Epigenetics. 2023 Dec;18(1):2282323. doi: 10.1080/15592294.2023.2282323. Epub 2023 Nov 27. Epigenetics. 2023. PMID: 38010265 Free PMC article.
-
Integration of Morphometrics and Machine Learning Enables Accurate Distinction between Wild and Farmed Common Carp.Life (Basel). 2022 Jun 25;12(7):957. doi: 10.3390/life12070957. Life (Basel). 2022. PMID: 35888047 Free PMC article.
-
Power Play of Commensal Bacteria in the Buccal Cavity of Female Nile Tilapia.Front Microbiol. 2021 Nov 16;12:773351. doi: 10.3389/fmicb.2021.773351. eCollection 2021. Front Microbiol. 2021. PMID: 34867911 Free PMC article.
-
Environment-driven reprogramming of gamete DNA methylation occurs during maturation and is transmitted intergenerationally in Atlantic Salmon.G3 (Bethesda). 2021 Dec 8;11(12):jkab353. doi: 10.1093/g3journal/jkab353. G3 (Bethesda). 2021. PMID: 34849830 Free PMC article.
References
-
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. - PubMed
-
- Larson G, Fuller DQ.. The evolution of animal domestication. Annu Rev Ecol Evol Syst. 2014;45:115–136.
-
- Roberge C, Einum S, Guderley H, et al. Rapid parallel evolutionary changes of gene transcription profiles in farmed Atlantic salmon. Mol Ecol. 2006;15:9–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
