A high-quality genome provides insights into the new taxonomic status and genomic characteristics of Cladopus chinensis (Podostemaceae)
- PMID: 32257232
- PMCID: PMC7109043
- DOI: 10.1038/s41438-020-0269-5
A high-quality genome provides insights into the new taxonomic status and genomic characteristics of Cladopus chinensis (Podostemaceae)
Abstract
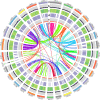
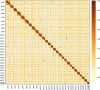
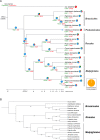
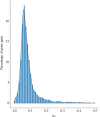
The Podostemaceae are ecologically and morphologically unusual aquatic angiosperms that survive only in rivers with pristine hydrology and high water quality and are at a relatively high risk of extinction. The taxonomic status of Podostemaceae has always been controversial. Here, we report the first high-quality genome assembly for Cladopus chinensis of Podostemaceae, obtained by incorporating Hi-C, Illumina and PacBio sequencing. We generated an 827.92 Mb genome with a contig N50 of 1.42 Mb and 27,370 annotated protein-coding genes. The assembled genome size was close to the estimated size, and 659.42 Mb of the assembly was assigned to 29 superscaffolds (scaffold N50 21.22 Mb). A total of 59.20% repetitive sequences were identified, among which long terminal repeats (LTRs) were the most abundant class (28.97% of the genome). Genome evolution analysis suggested that the divergence time of Cladopus chinensis (106 Mya) was earlier than that of Malpighiales (82 Mya) and that this taxon diverged into an independent branch of Podestemales. A recent whole-genome duplication (WGD) event occurred 4.43 million years ago. Comparative genomic analysis revealed that the expansion and contraction of oxidative phosphorylation, photosynthesis and isoflavonoid metabolism genes in Cladopus chinensis are probably related to the genomic characteristics of this growing submerged species. Transcriptome analysis revealed that upregulated genes in the shoot group compared to the root group were enriched in the NAC gene family and transcription factors associated with shoot development and defense responses, including WUSCHEL (WUS), ASYMMETRIC LEAVES (ASL), SHOOT MERISTEMLESS (STM), NAC2, NAC8, NAC29, NAC47, NAC73, NAC83 and NAC102. These findings provide new insights into the genomic diversity of unusual aquatic angiosperms and serve as a valuable reference for the taxonomic status and unusual shoot apical meristem of Podostemaceae.
Keywords: Bioinformatics; Genomic analysis; High-throughput screening.
© The Author(s) 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures



Similar articles
-
A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis].Hortic Res. 2020 Dec 28;7(1):212. doi: 10.1038/s41438-020-00449-z. Hortic Res. 2020. PMID: 33372175 Free PMC article.
-
Expression of SHOOT MERISTEMLESS, WUSCHEL, and ASYMMETRIC LEAVES1 homologs in the shoots of Podostemaceae: implications for the evolution of novel shoot organogenesis.Plant Cell. 2010 Jul;22(7):2131-40. doi: 10.1105/tpc.109.073189. Epub 2010 Jul 20. Plant Cell. 2010. PMID: 20647344 Free PMC article.
-
A high-quality chromosome-level genome of wild Rosa rugosa.DNA Res. 2021 Sep 13;28(5):dsab017. doi: 10.1093/dnares/dsab017. DNA Res. 2021. PMID: 34499118 Free PMC article.
-
Draft genome sequence of cauliflower (Brassica oleracea L. var. botrytis) provides new insights into the C genome in Brassica species.Hortic Res. 2019 Jul 1;6:82. doi: 10.1038/s41438-019-0164-0. eCollection 2019. Hortic Res. 2019. PMID: 31645943 Free PMC article.
-
Chromosome-level assembly of the mangrove plant Aegiceras corniculatum genome generated through Illumina, PacBio and Hi-C sequencing technologies.Mol Ecol Resour. 2021 Jul;21(5):1593-1607. doi: 10.1111/1755-0998.13347. Epub 2021 Mar 8. Mol Ecol Resour. 2021. PMID: 33550674
Cited by
-
Chromosome-scale assembly and population diversity analyses provide insights into the evolution of Sapindus mukorossi.Hortic Res. 2022 Jan 5;9:uhac012. doi: 10.1093/hr/uhac012. Hortic Res. 2022. PMID: 35178562 Free PMC article.
-
Elevated mutation rates underlie the evolution of the aquatic plant family Podostemaceae.Commun Biol. 2022 Jan 20;5(1):75. doi: 10.1038/s42003-022-03003-w. Commun Biol. 2022. PMID: 35058542 Free PMC article.
-
Chitosan Oligosaccharide Production Potential of Mitsuaria sp. C4 and Its Whole-Genome Sequencing.Front Microbiol. 2021 Aug 5;12:695571. doi: 10.3389/fmicb.2021.695571. eCollection 2021. Front Microbiol. 2021. PMID: 34421850 Free PMC article.
-
Comparative mitochondrial genomics of Terniopsis yongtaiensis in Malpighiales: structural, sequential, and phylogenetic perspectives.BMC Genomics. 2024 Sep 12;25(1):853. doi: 10.1186/s12864-024-10765-6. BMC Genomics. 2024. PMID: 39267005 Free PMC article.
-
Telomere-to-telomere and gap-free genome assembly of a susceptible grapevine species (Thompson Seedless) to facilitate grape functional genomics.Hortic Res. 2023 Dec 13;11(1):uhad260. doi: 10.1093/hr/uhad260. eCollection 2024 Jan. Hortic Res. 2023. PMID: 38288254 Free PMC article.
References
-
- Vinicius NL, et al. High importance of autochthonous basal food source for the food web of a brazilian tropical stream regardless of shading. Int. Rev. Hydrobiol. 2016;101:132–142. doi: 10.1002/iroh.201601851. - DOI
-
- Sobral-Leite M, et al. Anthecology and reproductive system of mourera fluviatilis (podostemaceae): pollination by bees and xenogamy in a predominantly anemophilous and autogamous family? Aquat. Bot. 2011;95:0–87. doi: 10.1016/j.aquabot.2011.03.010. - DOI
-
- Ram HYM, Sehgal A. In vitro studies on developmental morphology of indian podostemaceae. Aquat. Bot. 1997;57:0–132. doi: 10.1016/S0304-3770(96)01124-2. - DOI
-
- Koi S, Kato M. Paracladopus chanthaburiensis, a new species of Podostemaceae from thailand, with notes on its morphology, phylogeny and distribution. Taxon. 2008;57:201–210.
LinkOut - more resources
Full Text Sources
Miscellaneous

