Templated folding of intrinsically disordered proteins
- PMID: 32253236
- PMCID: PMC7212662
- DOI: 10.1074/jbc.REV120.012413
Templated folding of intrinsically disordered proteins
Abstract
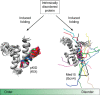
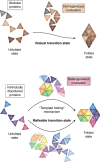
Much of our current knowledge of biological chemistry is founded in the structure-function relationship, whereby sequence determines structure that determines function. Thus, the discovery that a large fraction of the proteome is intrinsically disordered, while being functional, has revolutionized our understanding of proteins and raised new and interesting questions. Many intrinsically disordered proteins (IDPs) have been determined to undergo a disorder-to-order transition when recognizing their physiological partners, suggesting that their mechanisms of folding are intrinsically different from those observed in globular proteins. However, IDPs also follow some of the classic paradigms established for globular proteins, pointing to important similarities in their behavior. In this review, we compare and contrast the folding mechanisms of globular proteins with the emerging features of binding-induced folding of intrinsically disordered proteins. Specifically, whereas disorder-to-order transitions of intrinsically disordered proteins appear to follow rules of globular protein folding, such as the cooperative nature of the reaction, their folding pathways are remarkably more malleable, due to the heterogeneous nature of their folding nuclei, as probed by analysis of linear free-energy relationship plots. These insights have led to a new model for the disorder-to-order transition in IDPs termed "templated folding," whereby the binding partner dictates distinct structural transitions en route to product, while ensuring a cooperative folding.
Keywords: folding kinetics; intrinsically disordered protein; mutagenesis; protein chemistry; protein denaturation; protein folding; reaction mechanism; transition state.
© 2020 Toto et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
-
Folding and binding pathways of BH3-only proteins are encoded within their intrinsically disordered sequence, not templated by partner proteins.J Biol Chem. 2018 Jun 22;293(25):9718-9723. doi: 10.1074/jbc.RA118.002791. Epub 2018 May 1. J Biol Chem. 2018. PMID: 29716994 Free PMC article.
-
Molecular Recognition by Templated Folding of an Intrinsically Disordered Protein.Sci Rep. 2016 Feb 25;6:21994. doi: 10.1038/srep21994. Sci Rep. 2016. PMID: 26912067 Free PMC article.
-
Understanding the Binding Induced Folding of Intrinsically Disordered Proteins by Protein Engineering: Caveats and Pitfalls.Int J Mol Sci. 2020 May 15;21(10):3484. doi: 10.3390/ijms21103484. Int J Mol Sci. 2020. PMID: 32429036 Free PMC article. Review.
-
How Robust Is the Mechanism of Folding-Upon-Binding for an Intrinsically Disordered Protein?Biophys J. 2018 Apr 24;114(8):1889-1894. doi: 10.1016/j.bpj.2018.03.017. Biophys J. 2018. PMID: 29694866 Free PMC article.
Cited by
-
Phase separation modulates the assembly and dynamics of a polarity-related scaffold-signaling hub.Nat Commun. 2022 Nov 23;13(1):7181. doi: 10.1038/s41467-022-35000-2. Nat Commun. 2022. PMID: 36418326 Free PMC article.
-
The dynamic properties of a nuclear coactivator binding domain are evolutionarily conserved.Commun Biol. 2022 Mar 30;5(1):286. doi: 10.1038/s42003-022-03217-y. Commun Biol. 2022. PMID: 35354917 Free PMC article.
-
Fuzziness and Frustration in the Energy Landscape of Protein Folding, Function, and Assembly.Acc Chem Res. 2021 Mar 2;54(5):1251-1259. doi: 10.1021/acs.accounts.0c00813. Epub 2021 Feb 8. Acc Chem Res. 2021. PMID: 33550810 Free PMC article. Review.
-
Intrinsically Disordered Proteins: An Overview.Int J Mol Sci. 2022 Nov 14;23(22):14050. doi: 10.3390/ijms232214050. Int J Mol Sci. 2022. PMID: 36430530 Free PMC article. Review.
-
AI-Based Protein Interaction Screening and Identification (AISID).Int J Mol Sci. 2022 Oct 2;23(19):11685. doi: 10.3390/ijms231911685. Int J Mol Sci. 2022. PMID: 36232986 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

