CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection
- PMID: 32241893
- PMCID: PMC7183222
- DOI: 10.1073/pnas.2001165117
CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection
Abstract
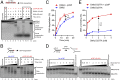
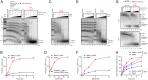
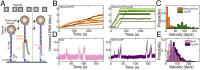
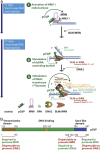
To repair a DNA double-strand break by homologous recombination, 5'-terminated DNA strands must first be resected to reveal 3'-overhangs. This process is initiated by a short-range resection catalyzed by MRE11-RAD50-NBS1 (MRN) stimulated by CtIP, which is followed by a long-range step involving EXO1 or DNA2 nuclease. DNA2 is a bifunctional enzyme that contains both single-stranded DNA (ssDNA)-specific nuclease and motor activities. Upon DNA unwinding by Bloom (BLM) or Werner (WRN) helicase, RPA directs the DNA2 nuclease to degrade the 5'-strand. RPA bound to ssDNA also represents a barrier, explaining the need for the motor activity of DNA2 to displace RPA prior to resection. Using ensemble and single-molecule biochemistry, we show that CtIP also dramatically stimulates the adenosine 5'-triphosphate (ATP) hydrolysis-driven motor activity of DNA2 involved in the long-range resection step. This activation in turn strongly promotes the degradation of RPA-coated ssDNA by DNA2. Accordingly, the stimulatory effect of CtIP is only observed with wild-type DNA2, but not the helicase-deficient variant. Similarly to the function of CtIP to promote MRN, also the DNA2 stimulatory effect is facilitated by CtIP phosphorylation. The domain of CtIP required to promote DNA2 is located in the central region lacking in lower eukaryotes and is fully separable from domains involved in the stimulation of MRN. These results establish how CtIP couples both MRE11-dependent short-range and DNA2-dependent long-range resection and define the involvement of the motor activity of DNA2 in this process. Our data might help explain the less severe resection defects of MRE11 nuclease-deficient cells compared to those lacking CtIP.
Keywords: DNA; DNA end resection; helicase; homologous recombination; nuclease.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Methods to Study DNA End Resection II: Biochemical Reconstitution Assays.Methods Enzymol. 2018;600:67-106. doi: 10.1016/bs.mie.2017.11.009. Epub 2018 Jan 9. Methods Enzymol. 2018. PMID: 29458776
-
Methods to Study DNA End Resection I: Recombinant Protein Purification.Methods Enzymol. 2018;600:25-66. doi: 10.1016/bs.mie.2017.11.008. Epub 2018 Feb 1. Methods Enzymol. 2018. PMID: 29458761
-
BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair.Genes Dev. 2011 Feb 15;25(4):350-62. doi: 10.1101/gad.2003811. Genes Dev. 2011. PMID: 21325134 Free PMC article.
-
The MRE11 complex: A versatile toolkit for the repair of broken DNA.DNA Repair (Amst). 2020 Jul-Aug;91-92:102869. doi: 10.1016/j.dnarep.2020.102869. Epub 2020 May 15. DNA Repair (Amst). 2020. PMID: 32480356 Review.
-
Mechanism and regulation of DNA end resection in eukaryotes.Crit Rev Biochem Mol Biol. 2016 May-Jun;51(3):195-212. doi: 10.3109/10409238.2016.1172552. Epub 2016 Apr 20. Crit Rev Biochem Mol Biol. 2016. PMID: 27098756 Free PMC article. Review.
Cited by
-
The CDK1-TOPBP1-PLK1 axis regulates the Bloom's syndrome helicase BLM to suppress crossover recombination in somatic cells.Sci Adv. 2022 Feb 4;8(5):eabk0221. doi: 10.1126/sciadv.abk0221. Epub 2022 Feb 4. Sci Adv. 2022. PMID: 35119917 Free PMC article.
-
Deciphering the mechanism of processive ssDNA digestion by the Dna2-RPA ensemble.Nat Commun. 2022 Jan 18;13(1):359. doi: 10.1038/s41467-021-27940-y. Nat Commun. 2022. PMID: 35042867 Free PMC article.
-
Homologous recombination deficiency: how genomic signatures are generated.Curr Opin Genet Dev. 2021 Feb;66:93-100. doi: 10.1016/j.gde.2021.01.002. Epub 2021 Jan 18. Curr Opin Genet Dev. 2021. PMID: 33477018 Free PMC article. Review.
-
The high toxicity of DSB-clusters modelling high-LET-DNA damage derives from inhibition of c-NHEJ and promotion of alt-EJ and SSA despite increases in HR.Front Cell Dev Biol. 2022 Oct 3;10:1016951. doi: 10.3389/fcell.2022.1016951. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36263011 Free PMC article.
-
Phosphorylated CtIP bridges DNA to promote annealing of broken ends.Proc Natl Acad Sci U S A. 2020 Sep 1;117(35):21403-21412. doi: 10.1073/pnas.2008645117. Epub 2020 Aug 19. Proc Natl Acad Sci U S A. 2020. PMID: 32817418 Free PMC article.
References
-
- Ranjha L., Howard S. M., Cejka P., Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 127, 187–214 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

