The hibernating 100S complex is a target of ribosome-recycling factor and elongation factor G in Staphylococcus aureus
- PMID: 32209660
- PMCID: PMC7196661
- DOI: 10.1074/jbc.RA119.012307
The hibernating 100S complex is a target of ribosome-recycling factor and elongation factor G in Staphylococcus aureus
Abstract
The formation of translationally inactive 70S dimers (called 100S ribosomes) by hibernation-promoting factor is a widespread survival strategy among bacteria. Ribosome dimerization is thought to be reversible, with the dissociation of the 100S complexes enabling ribosome recycling for participation in new rounds of translation. The precise pathway of 100S ribosome recycling has been unclear. We previously found that the heat-shock GTPase HflX in the human pathogen Staphylococcus aureus is a minor disassembly factor. Cells lacking hflX do not accumulate 100S ribosomes unless they are subjected to heat exposure, suggesting the existence of an alternative pathway during nonstressed conditions. Here, we provide biochemical and genetic evidence that two essential translation factors, ribosome-recycling factor (RRF) and GTPase elongation factor G (EF-G), synergistically split 100S ribosomes in a GTP-dependent but tRNA translocation-independent manner. We found that although HflX and the RRF/EF-G pair are functionally interchangeable, HflX is expressed at low levels and is dispensable under normal growth conditions. The bacterial RRF/EF-G pair was previously known to target only the post-termination 70S complexes; our results reveal a new role in the reversal of ribosome hibernation that is intimately linked to bacterial pathogenesis, persister formation, stress responses, and ribosome integrity.
Keywords: 100S ribosome; GTPase; Staphylococcus aureus (S. aureus); bacterial persistence; elongation factor G (EF-G); hibernation; ribosome; ribosome-recycling factor (RRF); translation elongation factor; translation regulation.
© 2020 Basu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



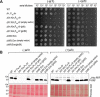
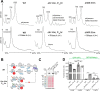

Similar articles
-
Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase.Proc Natl Acad Sci U S A. 2017 Sep 26;114(39):E8165-E8173. doi: 10.1073/pnas.1709588114. Epub 2017 Sep 11. Proc Natl Acad Sci U S A. 2017. PMID: 28894000 Free PMC article.
-
Thermal and Nutritional Regulation of Ribosome Hibernation in Staphylococcus aureus.J Bacteriol. 2018 Nov 26;200(24):e00426-18. doi: 10.1128/JB.00426-18. Print 2018 Dec 15. J Bacteriol. 2018. PMID: 30297357 Free PMC article.
-
The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G).Nucleic Acids Res. 2008 Dec;36(21):6676-87. doi: 10.1093/nar/gkn647. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948280 Free PMC article.
-
Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors.Biochimie. 1996;78(11-12):959-69. doi: 10.1016/s0300-9084(97)86718-1. Biochimie. 1996. PMID: 9150873 Review.
-
Mechanisms of ribosome recycling in bacteria and mitochondria: a structural perspective.RNA Biol. 2022;19(1):662-677. doi: 10.1080/15476286.2022.2067712. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35485608 Free PMC article. Review.
Cited by
-
Staphylococcal exoribonuclease YhaM destabilizes ribosomes by targeting the mRNA of a hibernation factor.Nucleic Acids Res. 2024 Aug 27;52(15):8998-9013. doi: 10.1093/nar/gkae596. Nucleic Acids Res. 2024. PMID: 38979572 Free PMC article.
-
Transcription regulates ribosome hibernation.Mol Microbiol. 2021 Aug;116(2):663-673. doi: 10.1111/mmi.14762. Epub 2021 Jun 21. Mol Microbiol. 2021. PMID: 34152658 Free PMC article.
-
Transposon sequencing identifies genes impacting Staphylococcus aureus invasion in a human macrophage model.Infect Immun. 2023 Oct 17;91(10):e0022823. doi: 10.1128/iai.00228-23. Epub 2023 Sep 7. Infect Immun. 2023. PMID: 37676013 Free PMC article.
-
Epitranscriptional m6A modification of rRNA negatively impacts translation and host colonization in Staphylococcus aureus.PLoS Pathog. 2024 Jan 22;20(1):e1011968. doi: 10.1371/journal.ppat.1011968. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38252661 Free PMC article.
-
The 27th Annual Midwest Microbial Pathogenesis Conference in the Age of COVID.J Bacteriol. 2022 Jun 21;204(6):e0013622. doi: 10.1128/jb.00136-22. Epub 2022 May 4. J Bacteriol. 2022. PMID: 35506693 Free PMC article. Review.
References
-
- Akiyama T., Williamson K. S., Schaefer R., Pratt S., Chang C. B., and Franklin M. J. (2017) Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc. Natl. Acad. Sci. U.S.A. 114, 3204–3209 10.1073/pnas.1700695114 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

