Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma
- PMID: 32203415
- PMCID: PMC7610258
- DOI: 10.1038/s41556-020-0481-4
Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma
Abstract
Although the transition metal copper (Cu) is an essential nutrient that is conventionally viewed as a static cofactor within enzyme active sites, a non-traditional role for Cu as a modulator of kinase signalling is emerging. Here, we found that Cu is required for the activity of the autophagic kinases ULK1 and ULK2 (ULK1/2) through a direct Cu-ULK1/2 interaction. Genetic loss of the Cu transporter Ctr1 or mutations in ULK1 that disrupt the binding of Cu reduced ULK1/2-dependent signalling and the formation of autophagosome complexes. Increased levels of intracellular Cu are associated with starvation-induced autophagy and are sufficient to enhance ULK1 kinase activity and, in turn, autophagic flux. The growth and survival of lung tumours driven by KRASG12D is diminished in the absence of Ctr1, is dependent on ULK1 Cu binding and is associated with reduced levels of autophagy and signalling. These findings suggest a molecular basis for exploiting Cu-chelation therapy to prevent autophagy signalling to limit proliferation and improve patient survival in cancer.
Conflict of interest statement
Figures



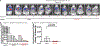
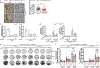

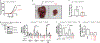

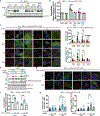

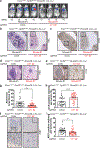

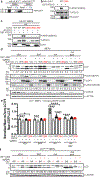
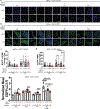
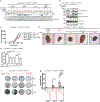
Comment in
-
Copper - a novel stimulator of autophagy.Cell Stress. 2020 Apr 24;4(5):92-94. doi: 10.15698/cst2020.05.218. Cell Stress. 2020. PMID: 32420529 Free PMC article.
Similar articles
-
ER-Targeted Beclin 1 Supports Autophagosome Biogenesis in the Absence of ULK1 and ULK2 Kinases.Cells. 2019 May 17;8(5):475. doi: 10.3390/cells8050475. Cells. 2019. PMID: 31108943 Free PMC article.
-
BRAFV600E-Driven Lung Adenocarcinoma Requires Copper to Sustain Autophagic Signaling and Processing.Mol Cancer Res. 2022 Jul 6;20(7):1096-1107. doi: 10.1158/1541-7786.MCR-21-0250. Mol Cancer Res. 2022. PMID: 35320362 Free PMC article.
-
AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms.Mol Cell Biol. 2018 Apr 30;38(10):e00023-18. doi: 10.1128/MCB.00023-18. Print 2018 May 15. Mol Cell Biol. 2018. PMID: 29507183 Free PMC article.
-
Function and regulation of ULK1: From physiology to pathology.Gene. 2022 Oct 5;840:146772. doi: 10.1016/j.gene.2022.146772. Epub 2022 Jul 26. Gene. 2022. PMID: 35905845 Review.
-
Mechanisms of Autophagy Initiation.Annu Rev Biochem. 2017 Jun 20;86:225-244. doi: 10.1146/annurev-biochem-061516-044820. Epub 2017 Mar 15. Annu Rev Biochem. 2017. PMID: 28301741 Free PMC article. Review.
Cited by
-
Cuproptosis patterns in papillary renal cell carcinoma are characterized by distinct tumor microenvironment infiltration landscapes.Front Mol Biosci. 2022 Oct 5;9:910928. doi: 10.3389/fmolb.2022.910928. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275614 Free PMC article.
-
Copper metabolism as a unique vulnerability in cancer.Biochim Biophys Acta Mol Cell Res. 2021 Feb;1868(2):118893. doi: 10.1016/j.bbamcr.2020.118893. Epub 2020 Oct 20. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33091507 Free PMC article. Review.
-
A novel cuproptosis-related molecular pattern and its tumor microenvironment characterization in colorectal cancer.Front Immunol. 2022 Sep 30;13:940774. doi: 10.3389/fimmu.2022.940774. eCollection 2022. Front Immunol. 2022. PMID: 36248908 Free PMC article.
-
Oxidation state-specific fluorescent copper sensors reveal oncogene-driven redox changes that regulate labile copper(II) pools.Proc Natl Acad Sci U S A. 2022 Oct 25;119(43):e2202736119. doi: 10.1073/pnas.2202736119. Epub 2022 Oct 17. Proc Natl Acad Sci U S A. 2022. PMID: 36252013 Free PMC article.
-
Cuproptosis-related lncRNA signatures: Predicting prognosis and evaluating the tumor immune microenvironment in lung adenocarcinoma.Front Oncol. 2023 Jan 17;12:1088931. doi: 10.3389/fonc.2022.1088931. eCollection 2022. Front Oncol. 2023. PMID: 36733364 Free PMC article.
References
-
- Kolch W, Halasz M, Granovskaya M & Kholodenko BN The dynamic control of signal transduction networks in cancer cells. Nat. Rev. Cancer 15, 515–527 (2015). - PubMed
-
- Chelly J et al. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat. Genet 3, 14–19 (1993). - PubMed
-
- Mercer JFB et al. Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat. Genet 3, 20–25 (1993). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RO1CA222503/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- R01 CA193603/CA/NCI NIH HHS/United States
- R35GM124749/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R35 GM124749/GM/NIGMS NIH HHS/United States
- R01 CA222503/CA/NCI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- T32 ES019851/ES/NIEHS NIH HHS/United States
- F31 CA243294/CA/NCI NIH HHS/United States
- RO1CA193603/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- T32ES019851/U.S. Department of Health & Human Services | NIH | National Institute of Environmental Health Sciences (NIEHS)/International
- F31CA243294/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

