Description of Virulent Factors and Horizontal Gene Transfers of Keratitis-Associated Amoeba Acanthamoeba Triangularis by Genome Analysis
- PMID: 32188120
- PMCID: PMC7157575
- DOI: 10.3390/pathogens9030217
Description of Virulent Factors and Horizontal Gene Transfers of Keratitis-Associated Amoeba Acanthamoeba Triangularis by Genome Analysis
Abstract
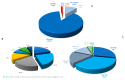
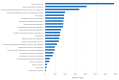
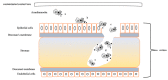
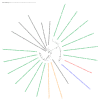
Acanthamoeba triangularis strain SH 621 is a free-living amoeba belonging to Acanthamoeba ribo-genotype T4. This ubiquitous protist is among the free-living amoebas responsible for Acanthamoeba keratitis, a severe infection of human cornea. Genome sequencing and genomic comparison were carried out to explore the biological functions and to better understand the virulence mechanism related to the pathogenicity of Acanthamoeba keratitis. The genome assembly harbored a length of 66.43 Mb encompassing 13,849 scaffolds. The analysis of predicted proteins reported the presence of 37,062 ORFs. A complete annotation revealed 33,168 and 16,605 genes that matched with NCBI non-redundant protein sequence (nr) and Cluster of Orthologous Group of proteins (COG) databases, respectively. The Kyoto Encyclopedia of Genes and Genomes Pathway (KEGG) annotation reported a great number of genes related to carbohydrate, amino acid and lipid metabolic pathways. The pangenome performed with 8 available amoeba genomes belonging to genus Acanthamoeba revealed a core genome containing 843 clusters of orthologous genes with a ratio core genome/pangenome of less than 0.02. We detected 48 genes related to virulent factors of Acanthamoeba keratitis. Best hit analyses in nr database identified 99 homologous genes shared with amoeba-resisting microorganisms. This study allows the deciphering the genome of a free-living amoeba with medical interest and provides genomic data to better understand virulence-related Acanthamoeba keratitis.
Keywords: Acanthamoeba triangularis; free-living amoebae; genes; genome; keratitis; pathogenicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A Comparative Genomic Approach to Determine the Virulence Factors and Horizontal Gene Transfer Events of Clinical Acanthamoeba Isolates.Microbiol Spectr. 2022 Apr 27;10(2):e0002522. doi: 10.1128/spectrum.00025-22. Epub 2022 Apr 13. Microbiol Spectr. 2022. PMID: 35416714 Free PMC article.
-
An Insight into the Genome of Pathogenic and Non-Pathogenic Acanthamoeba.Pathogens. 2022 Dec 19;11(12):1558. doi: 10.3390/pathogens11121558. Pathogens. 2022. PMID: 36558892 Free PMC article.
-
Genotypic characterization of Acanthamoeba spp. causing ocular infections in Swedish patients: identification of the T15 genotype in a case of protracted keratitis.Scand J Infect Dis. 2010 Oct;42(10):781-6. doi: 10.3109/00365548.2010.490563. Scand J Infect Dis. 2010. PMID: 20524785
-
Therapeutic agents and biocides for ocular infections by free-living amoebae of Acanthamoeba genus.Surv Ophthalmol. 2017 Mar-Apr;62(2):203-218. doi: 10.1016/j.survophthal.2016.10.009. Epub 2016 Nov 9. Surv Ophthalmol. 2017. PMID: 27836717 Review.
-
[Amoebas of the genus acanthamoeba as an etiological factor of keratitis].Klin Oczna. 2010;112(4-6):161-4. Klin Oczna. 2010. PMID: 20825074 Review. Polish.
Cited by
-
Distribution and Current State of Molecular Genetic Characterization in Pathogenic Free-Living Amoebae.Pathogens. 2022 Oct 18;11(10):1199. doi: 10.3390/pathogens11101199. Pathogens. 2022. PMID: 36297255 Free PMC article. Review.
-
Insight into the Lifestyle of Amoeba Willaertia magna during Bioreactor Growth Using Transcriptomics and Proteomics.Microorganisms. 2020 May 21;8(5):771. doi: 10.3390/microorganisms8050771. Microorganisms. 2020. PMID: 32455615 Free PMC article.
-
Peganum harmala Extract Has Antiamoebic Activity to Acanthamoeba triangularis Trophozoites and Changes Expression of Autophagy-Related Genes.Pathogens. 2021 Jul 4;10(7):842. doi: 10.3390/pathogens10070842. Pathogens. 2021. PMID: 34357992 Free PMC article.
-
Amoebicidal activity of Cassia angustifolia extract and its effect on Acanthamoeba triangularis autophagy-related gene expression at the transcriptional level.Parasitology. 2021 Aug;148(9):1074-1082. doi: 10.1017/S0031182021000718. Epub 2021 May 10. Parasitology. 2021. PMID: 33966667 Free PMC article.
-
A Comparative Genomic Approach to Determine the Virulence Factors and Horizontal Gene Transfer Events of Clinical Acanthamoeba Isolates.Microbiol Spectr. 2022 Apr 27;10(2):e0002522. doi: 10.1128/spectrum.00025-22. Epub 2022 Apr 13. Microbiol Spectr. 2022. PMID: 35416714 Free PMC article.
References
-
- Kot K., Łanocha-Arendarczyk N.A., Kosik-Bogacka D.I. Amoebas from the genus Acanthamoeba and their pathogenic properties. Ann. Parasitol. 2018;64:299–308. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

