SUNny Ways: The Role of the SUN-Domain Protein Mps3 Bridging Yeast Nuclear Organization and Lipid Homeostasis
- PMID: 32184804
- PMCID: PMC7058695
- DOI: 10.3389/fgene.2020.00136
SUNny Ways: The Role of the SUN-Domain Protein Mps3 Bridging Yeast Nuclear Organization and Lipid Homeostasis
Abstract
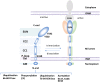
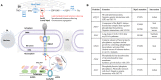
Mps3 is a SUN (Sad1-UNC-84) domain-containing protein that is located in the inner nuclear membrane (INM). Genetic screens with multiple Mps3 mutants have suggested that distinct regions of Mps3 function in relative isolation and underscore the broad involvement of Mps3 in multiple pathways including mitotic spindle formation, telomere maintenance, and lipid metabolism. These pathways have largely been characterized in isolation, without a holistic consideration for how key regulatory events within one pathway might impinge on other aspects of biology at the nuclear membrane. Mps3 is uniquely positioned to function in these multiple pathways as its N- terminus is in the nucleoplasm, where it is important for telomere anchoring at the nuclear periphery, and its C-terminus is in the lumen, where it has links with lipid metabolic processes. Emerging work suggests that the role of Mps3 in nuclear organization and lipid homeostasis are not independent, but more connected. For example, a failure in regulating Mps3 levels through the cell cycle leads to nuclear morphological abnormalities and loss of viability, suggesting a link between the N-terminal domain of Mps3 and nuclear envelope homeostasis. We will highlight work suggesting that Mps3 is pivotal factor in communicating events between the nucleus and the lipid bilayer.
Keywords: SUN-domain proteins; lipid metabolism; nuclear envelope; telomeres; transcription.
Copyright © 2020 Sosa Ponce, Moradi-Fard, Zaremberg and Cobb.
Figures
Similar articles
-
Genetic analysis of Mps3 SUN domain mutants in Saccharomyces cerevisiae reveals an interaction with the SUN-like protein Slp1.G3 (Bethesda). 2012 Dec;2(12):1703-18. doi: 10.1534/g3.112.004614. Epub 2012 Dec 1. G3 (Bethesda). 2012. PMID: 23275891 Free PMC article.
-
Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3.J Cell Biol. 2007 Dec 3;179(5):845-54. doi: 10.1083/jcb.200706040. Epub 2007 Nov 26. J Cell Biol. 2007. PMID: 18039933 Free PMC article.
-
Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z.J Cell Biol. 2011 May 2;193(3):489-507. doi: 10.1083/jcb.201011017. Epub 2011 Apr 25. J Cell Biol. 2011. PMID: 21518795 Free PMC article.
-
A genetic approach to study the role of nuclear envelope components in nuclear positioning.Novartis Found Symp. 2005;264:208-19; discussion 219-230. Novartis Found Symp. 2005. PMID: 15773756 Review.
-
Patrolling the nucleus: inner nuclear membrane-associated degradation.Curr Genet. 2019 Oct;65(5):1099-1106. doi: 10.1007/s00294-019-00971-1. Epub 2019 Apr 24. Curr Genet. 2019. PMID: 31020383 Free PMC article. Review.
Cited by
-
Nuclear mechanosignaling in striated muscle diseases.Front Physiol. 2023 Mar 7;14:1126111. doi: 10.3389/fphys.2023.1126111. eCollection 2023. Front Physiol. 2023. PMID: 36960155 Free PMC article. Review.
-
Phosphorylation of luminal region of the SUN-domain protein Mps3 promotes nuclear envelope localization during meiosis.Elife. 2021 Sep 29;10:e63119. doi: 10.7554/eLife.63119. Elife. 2021. PMID: 34586062 Free PMC article.
-
A Yeast Mitotic Tale for the Nucleus and the Vacuoles to Embrace.Int J Mol Sci. 2023 Jun 6;24(12):9829. doi: 10.3390/ijms24129829. Int J Mol Sci. 2023. PMID: 37372977 Free PMC article. Review.
-
Tuning between Nuclear Organization and Functionality in Health and Disease.Cells. 2023 Feb 23;12(5):706. doi: 10.3390/cells12050706. Cells. 2023. PMID: 36899842 Free PMC article. Review.
-
Homologous recombination within repetitive DNA.Curr Opin Genet Dev. 2021 Dec;71:143-153. doi: 10.1016/j.gde.2021.08.005. Epub 2021 Aug 28. Curr Opin Genet Dev. 2021. PMID: 34464817 Free PMC article. Review.
References
-
- Addinall S. G., Downey M., Yu M., Zubko M. K., Dewar J., Leake A., et al. (2008). A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180, 2251–2266. 10.1534/genetics.108.092577 - DOI - PMC - PubMed
-
- Addinall S. G., Holstein E. M., Lawless C., Yu M., Chapman K., Banks A. P., et al. (2011). Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PloS Genet. 7, e1001362. 10.1371/journal.pgen.1001362 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

