Kin28 depletion increases association of TFIID subunits Taf1 and Taf4 with promoters in Saccharomyces cerevisiae
- PMID: 32182349
- PMCID: PMC7192586
- DOI: 10.1093/nar/gkaa165
Kin28 depletion increases association of TFIID subunits Taf1 and Taf4 with promoters in Saccharomyces cerevisiae
Abstract
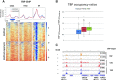
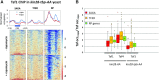
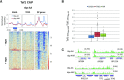
Transcription of eukaryotic mRNA-encoding genes by RNA polymerase II (Pol II) begins with assembly of the pre-initiation complex (PIC), comprising Pol II and the general transcription factors. Although the pathway of PIC assembly is well established, the mechanism of assembly and the dynamics of PIC components are not fully understood. For example, only recently has it been shown that in yeast, the Mediator complex normally occupies promoters only transiently, but shows increased association when Pol II promoter escape is inhibited. Here we show that two subunits of TFIID, Taf1 and Taf4, similarly show increased occupancy as measured by ChIP upon depletion or inactivation of Kin28. In contrast, TBP occupancy is unaffected by depletion of Kin28, thus revealing an uncoupling of Taf and TBP occupancy during the transcription cycle. Increased Taf1 occupancy upon Kin28 depletion is suppressed by depletion of TBP, while depletion of TBP in the presence of Kin28 has little effect on Taf1 occupancy. The increase in Taf occupancy upon depletion of Kin28 is more pronounced at TFIID-dominated promoters compared to SAGA-dominated promoters. Our results support the suggestion, based on recent structural studies, that TFIID may not remain bound to gene promoters through the transcription initiation cycle.
Published by Oxford University Press on behalf of Nucleic Acids Research 2020.
Figures



Similar articles
-
SAGA mediates transcription from the TATA-like element independently of Taf1p/TFIID but dependent on core promoter structures in Saccharomyces cerevisiae.PLoS One. 2017 Nov 27;12(11):e0188435. doi: 10.1371/journal.pone.0188435. eCollection 2017. PLoS One. 2017. PMID: 29176831 Free PMC article.
-
Mechanistic Differences in Transcription Initiation at TATA-Less and TATA-Containing Promoters.Mol Cell Biol. 2017 Dec 13;38(1):e00448-17. doi: 10.1128/MCB.00448-17. Print 2018 Jan 1. Mol Cell Biol. 2017. PMID: 29038161 Free PMC article.
-
Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II.Nucleic Acids Res. 2008 Mar;36(4):1343-57. doi: 10.1093/nar/gkm1068. Epub 2008 Jan 10. Nucleic Acids Res. 2008. PMID: 18187511 Free PMC article.
-
Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription.Gene. 2003 Oct 2;315:1-13. doi: 10.1016/s0378-1119(03)00714-5. Gene. 2003. PMID: 14557059 Review.
-
SAGA and TFIID: Friends of TBP drifting apart.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194604. doi: 10.1016/j.bbagrm.2020.194604. Epub 2020 Jul 14. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32673655 Review.
Cited by
-
An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters.Genes Dev. 2022 Sep 1;36(17-18):985-1001. doi: 10.1101/gad.350026.122. Epub 2022 Oct 27. Genes Dev. 2022. PMID: 36302553 Free PMC article.
-
Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction.Elife. 2020 Apr 27;9:e55667. doi: 10.7554/eLife.55667. Elife. 2020. PMID: 32338606 Free PMC article.
-
Spatiotemporal coordination of transcription preinitiation complex assembly in live cells.Mol Cell. 2021 Sep 2;81(17):3560-3575.e6. doi: 10.1016/j.molcel.2021.07.022. Epub 2021 Aug 9. Mol Cell. 2021. PMID: 34375585 Free PMC article.
-
Mapping quantitative trait loci and developing their KASP markers for pre-harvest sprouting resistance of Henan wheat varieties in China.Front Plant Sci. 2023 Feb 2;14:1118777. doi: 10.3389/fpls.2023.1118777. eCollection 2023. Front Plant Sci. 2023. PMID: 36875573 Free PMC article.
-
A mitogen-activated protein kinase PoxMK1 mediates regulation of the production of plant-biomass-degrading enzymes, vegetative growth, and pigment biosynthesis in Penicillium oxalicum.Appl Microbiol Biotechnol. 2021 Jan;105(2):661-678. doi: 10.1007/s00253-020-11020-0. Epub 2021 Jan 6. Appl Microbiol Biotechnol. 2021. PMID: 33409610
References
-
- Sainsbury S., Bernecky C., Cramer P.. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:129–143. - PubMed
-
- Eisenmann D.M., Arndt K.M., Ricupero S.L., Rooney J.W., Winston F.. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992; 6:1319–1331. - PubMed
-
- Huisinga K.L., Pugh B.F.. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004; 13:573–585. - PubMed
-
- Kuras L., Kosa P., Mencia M., Struhl K.. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000; 288:1244–1248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

