Ldb1 is required for Lmo2 oncogene-induced thymocyte self-renewal and T-cell acute lymphoblastic leukemia
- PMID: 32181817
- PMCID: PMC7316212
- DOI: 10.1182/blood.2019000794
Ldb1 is required for Lmo2 oncogene-induced thymocyte self-renewal and T-cell acute lymphoblastic leukemia
Abstract
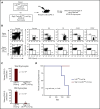
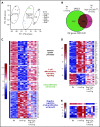
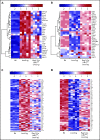
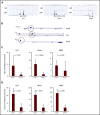
Prolonged or enhanced expression of the proto-oncogene Lmo2 is associated with a severe form of T-cell acute lymphoblastic leukemia (T-ALL), designated early T-cell precursor ALL, which is characterized by the aberrant self-renewal and subsequent oncogenic transformation of immature thymocytes. It has been suggested that Lmo2 exerts these effects by functioning as component of a multi-subunit transcription complex that includes the ubiquitous adapter Ldb1 along with b-HLH and/or GATA family transcription factors; however, direct experimental evidence for this mechanism is lacking. In this study, we investigated the importance of Ldb1 for Lmo2-induced T-ALL by conditional deletion of Ldb1 in thymocytes in an Lmo2 transgenic mouse model of T-ALL. Our results identify a critical requirement for Ldb1 in Lmo2-induced thymocyte self-renewal and thymocyte radiation resistance and for the transition of preleukemic thymocytes to overt T-ALL. Moreover, Ldb1 was also required for acquisition of the aberrant preleukemic ETP gene expression signature in immature Lmo2 transgenic thymocytes. Co-binding of Ldb1 and Lmo2 was detected at the promoters of key upregulated T-ALL driver genes (Hhex, Lyl1, and Nfe2) in preleukemic Lmo2 transgenic thymocytes, and binding of both Ldb1 and Lmo2 at these sites was reduced following Cre-mediated deletion of Ldb1. Together, these results identify a key role for Ldb1, a nonproto-oncogene, in T-ALL and support a model in which Lmo2-induced T-ALL results from failure to downregulate Ldb1/Lmo2-nucleated transcription complexes which normally function to enforce self-renewal in bone marrow hematopoietic progenitors.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
Similar articles
-
Hhex regulates Kit to promote radioresistance of self-renewing thymocytes in Lmo2-transgenic mice.Leukemia. 2015 Apr;29(4):927-38. doi: 10.1038/leu.2014.292. Epub 2014 Oct 6. Leukemia. 2015. PMID: 25283843
-
Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL.Blood. 2013 Sep 19;122(12):2093-103. doi: 10.1182/blood-2012-09-458570. Epub 2013 Aug 7. Blood. 2013. PMID: 23926305
-
The NUP98-HOXD13 fusion oncogene induces thymocyte self-renewal via Lmo2/Lyl1.Leukemia. 2019 Aug;33(8):1868-1880. doi: 10.1038/s41375-018-0361-0. Epub 2019 Jan 30. Leukemia. 2019. PMID: 30700838
-
SCL/TAL1 in Hematopoiesis and Cellular Reprogramming.Curr Top Dev Biol. 2016;118:163-204. doi: 10.1016/bs.ctdb.2016.01.004. Epub 2016 Feb 18. Curr Top Dev Biol. 2016. PMID: 27137657 Review.
-
Thymocyte self-renewal and oncogenic risk in immunodeficient mouse models: relevance for human gene therapy clinical trials targeting haematopoietic stem cell populations?Mamm Genome. 2018 Dec;29(11-12):771-776. doi: 10.1007/s00335-018-9780-5. Epub 2018 Sep 4. Mamm Genome. 2018. PMID: 30182300 Review.
Cited by
-
A case of T-cell acute lymphoblastic leukemia in retroviral gene therapy for ADA-SCID.Nat Commun. 2024 Apr 30;15(1):3662. doi: 10.1038/s41467-024-47866-5. Nat Commun. 2024. PMID: 38688902 Free PMC article.
-
LMO2 promotes the development of AML through interaction with transcription co-regulator LDB1.Cell Death Dis. 2023 Aug 12;14(8):518. doi: 10.1038/s41419-023-06039-w. Cell Death Dis. 2023. PMID: 37573405 Free PMC article.
-
Potential Candidate Genes for Therapeutic Targeting in Chronic Myeloid Leukemia: A Pilot Study.Asian Pac J Cancer Prev. 2023 Sep 1;24(9):3077-3085. doi: 10.31557/APJCP.2023.24.9.3077. Asian Pac J Cancer Prev. 2023. PMID: 37774059 Free PMC article.
-
LDB1 Enforces Stability on Direct and Indirect Oncoprotein Partners in Leukemia.Mol Cell Biol. 2020 May 28;40(12):e00652-19. doi: 10.1128/MCB.00652-19. Print 2020 May 28. Mol Cell Biol. 2020. PMID: 32229578 Free PMC article.
-
Multi-modular structure of the gene regulatory network for specification and commitment of murine T cells.Front Immunol. 2023 Jan 31;14:1108368. doi: 10.3389/fimmu.2023.1108368. eCollection 2023. Front Immunol. 2023. PMID: 36817475 Free PMC article. Review.
References
-
- Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16(8):494-507. - PubMed
-
- Fisch P, Boehm T, Lavenir I, et al. . T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7(12):2389-2397. - PubMed
-
- Larson RC, Fisch P, Larson TA, et al. . T cell tumours of disparate phenotype in mice transgenic for Rbtn-2. Oncogene. 1994;9(12):3675-3681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

