Yeast as a Model to Understand Actin-Mediated Cellular Functions in Mammals-Illustrated with Four Actin Cytoskeleton Proteins
- PMID: 32164332
- PMCID: PMC7140605
- DOI: 10.3390/cells9030672
Yeast as a Model to Understand Actin-Mediated Cellular Functions in Mammals-Illustrated with Four Actin Cytoskeleton Proteins
Abstract
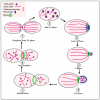
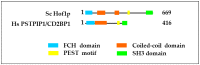
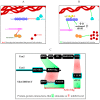
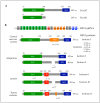
The budding yeast Saccharomyces cerevisiae has an actin cytoskeleton that comprises a set of protein components analogous to those found in the actin cytoskeletons of higher eukaryotes. Furthermore, the actin cytoskeletons of S. cerevisiae and of higher eukaryotes have some similar physiological roles. The genetic tractability of budding yeast and the availability of a stable haploid cell type facilitates the application of molecular genetic approaches to assign functions to the various actin cytoskeleton components. This has provided information that is in general complementary to that provided by studies of the equivalent proteins of higher eukaryotes and hence has enabled a more complete view of the role of these proteins. Several human functional homologues of yeast actin effectors are implicated in diseases. A better understanding of the molecular mechanisms underpinning the functions of these proteins is critical to develop improved therapeutic strategies. In this article we chose as examples four evolutionarily conserved proteins that associate with the actin cytoskeleton: 1) yeast Hof1p/mammalian PSTPIP1, 2) yeast Rvs167p/mammalian BIN1, 3) yeast eEF1A/eEF1A1 and eEF1A2 and 4) yeast Yih1p/mammalian IMPACT. We compare the knowledge on the functions of these actin cytoskeleton-associated proteins that has arisen from studies of their homologues in yeast with information that has been obtained from in vivo studies using live animals or in vitro studies using cultured animal cell lines.
Keywords: BAR domain; F-BAR domain; Myc; WASP; Wiskott-Aldrich Syndrome; cancer; cytokinesis; endocytosis; translation factors; tumor suppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The yeast actin cytoskeleton.FEMS Microbiol Rev. 2014 Mar;38(2):213-27. doi: 10.1111/1574-6976.12064. Epub 2014 Feb 20. FEMS Microbiol Rev. 2014. PMID: 24467403 Review.
-
The yeast actin cytoskeleton: from cellular function to biochemical mechanism.Microbiol Mol Biol Rev. 2006 Sep;70(3):605-45. doi: 10.1128/MMBR.00013-06. Microbiol Mol Biol Rev. 2006. PMID: 16959963 Free PMC article. Review.
-
Verprolin function in endocytosis and actin organization. Roles of the Las17p (yeast WASP)-binding domain and a novel C-terminal actin-binding domain.FEBS J. 2007 Aug;274(16):4103-25. doi: 10.1111/j.1742-4658.2007.05936.x. Epub 2007 Jul 16. FEBS J. 2007. PMID: 17635585
-
Las17p-Vrp1p but not Las17p-Arp2/3 interaction is important for actin patch polarization in yeast.Biochim Biophys Acta. 2009 May;1793(5):825-35. doi: 10.1016/j.bbamcr.2009.02.012. Epub 2009 Mar 9. Biochim Biophys Acta. 2009. PMID: 19272406
-
Yeast studies reveal moonlighting functions of the ancient actin cytoskeleton.IUBMB Life. 2014 Aug;66(8):538-45. doi: 10.1002/iub.1294. Epub 2014 Aug 19. IUBMB Life. 2014. PMID: 25138357 Free PMC article. Review.
Cited by
-
A census of actin-associated proteins in humans.Front Cell Dev Biol. 2023 Apr 28;11:1168050. doi: 10.3389/fcell.2023.1168050. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37187613 Free PMC article.
-
MicroRNA PC-3p-2869 Regulates Antler Growth and Inhibits Proliferation and Migration of Human Osteosarcoma and Chondrosarcoma Cells by Targeting CDK8, EEF1A1, and NTN1.Int J Mol Sci. 2023 Jun 29;24(13):10840. doi: 10.3390/ijms241310840. Int J Mol Sci. 2023. PMID: 37446017 Free PMC article.
-
Single-Locus and Multi-Locus Genome-Wide Association Studies Identify Genes Associated with Liver Cu Concentration in Merinoland Sheep.Genes (Basel). 2023 May 8;14(5):1053. doi: 10.3390/genes14051053. Genes (Basel). 2023. PMID: 37239413 Free PMC article.
-
Oceanapiside, a Marine Natural Product, Targets the Sphingolipid Pathway of Fluconazole-Resistant Candida glabrata.Mar Drugs. 2021 Feb 26;19(3):126. doi: 10.3390/md19030126. Mar Drugs. 2021. PMID: 33652774 Free PMC article.
-
Unravelling bacterial virulence factors in yeast: From identification to the elucidation of their mechanisms of action.Arch Microbiol. 2024 Jun 15;206(7):303. doi: 10.1007/s00203-024-04023-2. Arch Microbiol. 2024. PMID: 38878203 Review.
References
-
- Broach J.R., Pringle J.R., Jones E.W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1991. Volume 1 Genome Dynamics, Protein Synthesis, and Energetics.
-
- Jones E.W., Pringle J.R., Broach J.R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1992. Volume II Gene Expression.
-
- Pringle J.R., Broach J.R., Jones E.W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1997. Volume III Cell Cycle and Cell Biology.
-
- Guide to Yeast Genetics and Molecular Biology. Method Enzymol. 1991;194:1–863. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

