Zika virus non-structural protein NS4A restricts eye growth in Drosophila through regulation of JAK/STAT signaling
- PMID: 32152180
- PMCID: PMC7197722
- DOI: 10.1242/dmm.040816
Zika virus non-structural protein NS4A restricts eye growth in Drosophila through regulation of JAK/STAT signaling
Abstract
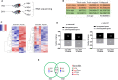
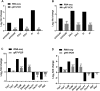
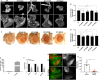
To gain a comprehensive view of the changes in host gene expression underlying Zika virus (ZIKV) pathogenesis, we performed whole-genome RNA sequencing (RNA-seq) of ZIKV-infected Drosophila adult flies. RNA-seq analysis revealed that ZIKV infection alters several and diverse biological processes, including stress, locomotion, lipid metabolism, imaginal disc morphogenesis and regulation of JAK/STAT signaling. To explore the interaction between ZIKV infection and JAK/STAT signaling regulation, we generated genetic constructs overexpressing ZIKV-specific non-structural proteins NS2A, NS2B, NS4A and NS4B. We found that ectopic expression of non-structural proteins in the developing Drosophila eye significantly restricts growth of the larval and adult eye and correlates with considerable repression of the in vivo JAK/STAT reporter, 10XStat92E-GFP At the cellular level, eye growth defects are associated with reduced rate of proliferation without affecting the overall rate of apoptosis. In addition, ZIKV NS4A genetically interacts with the JAK/STAT signaling components; co-expression of NS4A along with the dominant-negative form of domeless or StatRNAi results in aggravated reduction in eye size, while co-expression of NS4A in HopTuml (also known as hopTum ) mutant background partially rescues the hop-induced eye overgrowth phenotype. The function of ZIKV NS4A in regulating growth is maintained in the wing, where ZIKV NS4A overexpression in the pouch domain results in reduced growth linked with diminished expression of Notch targets, Wingless (Wg) and Cut, and the Notch reporter, NRE-GFP Thus, our study provides evidence that ZIKV infection in Drosophila results in restricted growth of the developing eye and wing, wherein eye phenotype is induced through regulation of JAK/STAT signaling, whereas restricted wing growth is induced through regulation of Notch signaling. The interaction of ZIKV non-structural proteins with the conserved host signaling pathways further advance our understanding of ZIKV-induced pathogenesis.This article has an associated First Person interview with the first author of the paper.
Keywords: Drosophila; Eye development; Host-pathogen interaction; JAK/STAT signaling; Zika virus.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
Similar articles
-
JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila.Development. 2006 Dec;133(23):4721-9. doi: 10.1242/dev.02675. Epub 2006 Nov 1. Development. 2006. PMID: 17079268
-
GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo.Gene Expr Patterns. 2007 Jan;7(3):323-31. doi: 10.1016/j.modgep.2006.08.003. Epub 2006 Aug 22. Gene Expr Patterns. 2007. PMID: 17008134
-
JAK/STAT signaling is required for hinge growth and patterning in the Drosophila wing disc.Dev Biol. 2013 Oct 15;382(2):413-26. doi: 10.1016/j.ydbio.2013.08.016. Epub 2013 Aug 23. Dev Biol. 2013. PMID: 23978534 Free PMC article.
-
Organogenesis and tumorigenesis: insight from the JAK/STAT pathway in the Drosophila eye.Dev Dyn. 2010 Oct;239(10):2522-33. doi: 10.1002/dvdy.22394. Dev Dyn. 2010. PMID: 20737505 Free PMC article. Review.
-
JAK/STAT pathway dysregulation in tumors: a Drosophila perspective.Semin Cell Dev Biol. 2014 Apr;28:96-103. doi: 10.1016/j.semcdb.2014.03.023. Epub 2014 Mar 28. Semin Cell Dev Biol. 2014. PMID: 24685611 Free PMC article. Review.
Cited by
-
Zika virus-induces metabolic alterations in fetal neuronal progenitors that could influence in neurodevelopment during early pregnancy.Biol Open. 2023 Apr 15;12(4):bio059889. doi: 10.1242/bio.059889. Epub 2023 Apr 21. Biol Open. 2023. PMID: 37093064 Free PMC article.
-
Flavivirus Infection and Regulation of Host Immune and Tissue Homeostasis in Insects.Front Immunol. 2020 Nov 30;11:618801. doi: 10.3389/fimmu.2020.618801. eCollection 2020. Front Immunol. 2020. PMID: 33329613 Free PMC article. Review. No abstract available.
-
Drosophila as a Model for Infectious Diseases.Int J Mol Sci. 2021 Mar 8;22(5):2724. doi: 10.3390/ijms22052724. Int J Mol Sci. 2021. PMID: 33800390 Free PMC article. Review.
-
The role of viral infection in implantation failure: direct and indirect effects.Reprod Biol Endocrinol. 2024 Nov 11;22(1):142. doi: 10.1186/s12958-024-01303-w. Reprod Biol Endocrinol. 2024. PMID: 39529140 Free PMC article. Review.
-
Host Factors That Control Mosquito-Borne Viral Infections in Humans and Their Vector.Viruses. 2021 Apr 24;13(5):748. doi: 10.3390/v13050748. Viruses. 2021. PMID: 33923307 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

