Transcriptomic signatures of brain regional vulnerability to Parkinson's disease
- PMID: 32139796
- PMCID: PMC7058608
- DOI: 10.1038/s42003-020-0804-9
Transcriptomic signatures of brain regional vulnerability to Parkinson's disease
Abstract
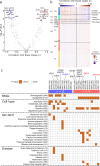
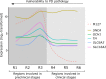
The molecular mechanisms underlying caudal-to-rostral progression of Lewy body pathology in Parkinson's disease remain poorly understood. Here, we identified transcriptomic signatures across brain regions involved in Braak Lewy body stages in non-neurological adults from the Allen Human Brain Atlas. Among the genes that are indicative of regional vulnerability, we found known genetic risk factors for Parkinson's disease: SCARB2, ELOVL7, SH3GL2, SNCA, BAP1, and ZNF184. Results were confirmed in two datasets of non-neurological subjects, while in two datasets of Parkinson's disease patients we found altered expression patterns. Co-expression analysis across vulnerable regions identified a module enriched for genes associated with dopamine synthesis and microglia, and another module related to the immune system, blood-oxygen transport, and endothelial cells. Both were highly expressed in regions involved in the preclinical stages of the disease. Finally, alterations in genes underlying these region-specific functions may contribute to the selective regional vulnerability in Parkinson's disease brains.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Transcriptomic profiling of Parkinson's disease brains reveals disease stage specific gene expression changes.Acta Neuropathol. 2023 Aug;146(2):227-244. doi: 10.1007/s00401-023-02597-7. Epub 2023 Jun 22. Acta Neuropathol. 2023. PMID: 37347276 Free PMC article.
-
Regional brain iron and gene expression provide insights into neurodegeneration in Parkinson's disease.Brain. 2021 Jul 28;144(6):1787-1798. doi: 10.1093/brain/awab084. Brain. 2021. PMID: 33704443 Free PMC article.
-
Review: Clinical, neuropathological and genetic features of Lewy body dementias.Neuropathol Appl Neurobiol. 2019 Dec;45(7):635-654. doi: 10.1111/nan.12554. Epub 2019 May 20. Neuropathol Appl Neurobiol. 2019. PMID: 30977926 Review.
-
Is Braak staging valid for all types of Parkinson's disease?J Neural Transm (Vienna). 2019 Apr;126(4):423-431. doi: 10.1007/s00702-018-1898-9. Epub 2018 Jun 25. J Neural Transm (Vienna). 2019. PMID: 29943229 Review.
-
In vivo microglia activation in very early dementia with Lewy bodies, comparison with Parkinson's disease.Parkinsonism Relat Disord. 2013 Jan;19(1):47-52. doi: 10.1016/j.parkreldis.2012.07.002. Epub 2012 Jul 26. Parkinsonism Relat Disord. 2013. PMID: 22841687
Cited by
-
Molecular characterization of the stress network in individuals at risk for schizophrenia.Neurobiol Stress. 2021 Feb 10;14:100307. doi: 10.1016/j.ynstr.2021.100307. eCollection 2021 May. Neurobiol Stress. 2021. PMID: 33644266 Free PMC article.
-
Variable Effects of PD-Risk Associated SNPs and Variants in Parkinsonism-Associated Genes on Disease Phenotype in a Community-Based Cohort.Front Neurol. 2021 Apr 14;12:662278. doi: 10.3389/fneur.2021.662278. eCollection 2021. Front Neurol. 2021. PMID: 33935957 Free PMC article.
-
Correcting Differential Gene Expression Analysis for Cyto-Architectural Alterations in Substantia Nigra of Parkinson's Disease Patients Reveals Known and Potential Novel Disease-Associated Genes and Pathways.Cells. 2022 Jan 7;11(2):198. doi: 10.3390/cells11020198. Cells. 2022. PMID: 35053314 Free PMC article.
-
Effects of the administration of Elovl5-dependent fatty acids on a spino-cerebellar ataxia 38 mouse model.Behav Brain Funct. 2022 Aug 6;18(1):8. doi: 10.1186/s12993-022-00194-4. Behav Brain Funct. 2022. PMID: 35933444 Free PMC article.
-
Interferon-γ signaling synergizes with LRRK2 in neurons and microglia derived from human induced pluripotent stem cells.Nat Commun. 2020 Oct 14;11(1):5163. doi: 10.1038/s41467-020-18755-4. Nat Commun. 2020. PMID: 33057020 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

