Plasmin improves blood-gas barrier function in oedematous lungs by cleaving epithelial sodium channels
- PMID: 32133621
- PMCID: PMC7280014
- DOI: 10.1111/bph.15038
Plasmin improves blood-gas barrier function in oedematous lungs by cleaving epithelial sodium channels
Abstract
Background and purpose: Lung oedema in association with suppressed fibrinolysis is a hallmark of lung injury. Here, we have tested whether plasmin cleaves epithelial sodium channels (ENaC) to resolve lung oedema fluid.
Experimental approach: Human lungs and airway acid-instilled mice were used for analysing fluid resolution. In silico prediction, mutagenesis, Xenopus oocytes, immunoblotting, voltage clamp, mass spectrometry, and protein docking were combined for identifying plasmin cleavage sites.
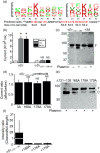
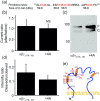
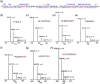
Key results: Plasmin improved lung fluid resolution in both human lungs ex vivo and injured mice. Plasmin activated αβγENaC channels in oocytes in a time-dependent manner. Deletion of four consensus proteolysis tracts (αΔ432-444, γΔ131-138, γΔ178-193, and γΔ410-422) eliminated plasmin-induced activation significantly. Further, immunoblotting assays identified 7 cleavage sites (K126, R135, K136, R153, K168, R178, K179) for plasmin to trim both furin-cleaved C-terminal fragments and full-length human γENaC proteins. In addition, 9 new sites (R122, R137, R138, K150, K170, R172, R180, K181, K189) in synthesized peptides were found to be cleaved by plasmin. These cleavage sites were located in the finger and the thumb, particularly the GRIP domain of human ENaC 3D model composed of two proteolytic centres for plasmin. Novel uncleaved sites beyond the GRIP domain in both α and γ subunits were identified to interrupt the plasmin cleavage-induced conformational change in ENaC channel complexes. Additionally, plasmin could regulate ENaC activity via the G protein signal.
Conclusion and implications: Plasmin can cleave ENaC to improve blood-gas exchange by resolving oedema fluid and could be a potent therapy for oedematous lungs.
© 2020 The British Pharmacological Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

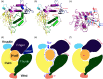
Similar articles
-
Plasmin and chymotrypsin have distinct preferences for channel activating cleavage sites in the γ subunit of the human epithelial sodium channel.J Gen Physiol. 2012 Oct;140(4):375-89. doi: 10.1085/jgp.201110763. Epub 2012 Sep 10. J Gen Physiol. 2012. PMID: 22966015 Free PMC article.
-
Plasmin activates epithelial Na+ channels by cleaving the gamma subunit.J Biol Chem. 2008 Dec 26;283(52):36586-91. doi: 10.1074/jbc.M805676200. Epub 2008 Nov 3. J Biol Chem. 2008. PMID: 18981180 Free PMC article.
-
Preferential assembly of epithelial sodium channel (ENaC) subunits in Xenopus oocytes: role of furin-mediated endogenous proteolysis.J Biol Chem. 2008 Mar 21;283(12):7455-63. doi: 10.1074/jbc.M707399200. Epub 2008 Jan 14. J Biol Chem. 2008. PMID: 18195015 Free PMC article.
-
Proteolytic Activation of the Epithelial Sodium Channel (ENaC): Its Mechanisms and Implications.Int J Mol Sci. 2023 Dec 16;24(24):17563. doi: 10.3390/ijms242417563. Int J Mol Sci. 2023. PMID: 38139392 Free PMC article. Review.
-
New role for plasmin in sodium homeostasis.Curr Opin Nephrol Hypertens. 2010 Jan;19(1):13-9. doi: 10.1097/MNH.0b013e3283330fb2. Curr Opin Nephrol Hypertens. 2010. PMID: 19864949 Free PMC article. Review.
Cited by
-
Fibrinolytic niche is required for alveolar type 2 cell-mediated alveologenesis via a uPA-A6-CD44+-ENaC signal cascade.Signal Transduct Target Ther. 2021 Feb 27;6(1):97. doi: 10.1038/s41392-021-00511-9. Signal Transduct Target Ther. 2021. PMID: 33640905 Free PMC article. No abstract available.
-
Functional prediction and comparative population analysis of variants in genes for proteases and innate immunity related to SARS-CoV-2 infection.Infect Genet Evol. 2020 Oct;84:104498. doi: 10.1016/j.meegid.2020.104498. Epub 2020 Aug 7. Infect Genet Evol. 2020. PMID: 32771700 Free PMC article.
-
Competitive cleavage of SARS-CoV-2 spike protein and epithelial sodium channel by plasmin as a potential mechanism for COVID-19 infection.Am J Physiol Lung Cell Mol Physiol. 2022 Nov 1;323(5):L569-L577. doi: 10.1152/ajplung.00152.2022. Epub 2022 Oct 4. Am J Physiol Lung Cell Mol Physiol. 2022. PMID: 36193902 Free PMC article.
-
Regulation of Epithelial Sodium Transport by SARS-CoV-2 Is Closely Related with Fibrinolytic System-Associated Proteins.Biomolecules. 2023 Mar 23;13(4):578. doi: 10.3390/biom13040578. Biomolecules. 2023. PMID: 37189326 Free PMC article. Review.
-
PAI-1 regulates AT2-mediated re-alveolarization and ion permeability.Res Sq [Preprint]. 2023 Mar 1:rs.3.rs-2289657. doi: 10.21203/rs.3.rs-2289657/v1. Res Sq. 2023. Update in: Stem Cell Res Ther. 2023 Jul 27;14(1):185. doi: 10.1186/s13287-023-03414-4. PMID: 36909505 Free PMC article. Updated. Preprint.
References
-
- Asakura, H. , Ontachi, Y. , Mizutani, T. , Kato, M. , Saito, M. , Kumabashiri, I. , … Nakao, S. (2001). An enhanced fibrinolysis prevents the development of multiple organ failure in disseminated intravascular coagulation in spite of much activation of blood coagulation. Critical Care Medicine, 29, 1164–1168. 10.1097/00003246-200106000-00015 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

