Characterization of human respiratory syncytial virus (RSV) isolated from HIV-exposed-uninfected and HIV-unexposed infants in South Africa during 2015-2017
- PMID: 32126161
- PMCID: PMC7298309
- DOI: 10.1111/irv.12727
Characterization of human respiratory syncytial virus (RSV) isolated from HIV-exposed-uninfected and HIV-unexposed infants in South Africa during 2015-2017
Abstract
Background: RSV is a leading cause of lower respiratory tract infection in infants. Monitoring RSV glycoprotein sequences is critical for understanding RSV epidemiology and viral antigenicity in the effort to develop anti-RSV prophylactics and therapeutics.
Objectives: The objective is to characterize the circulating RSV strains collected from infants in South Africa during 2015-2017.
Methods: A subset of 150 RSV-positive samples obtained in South Africa from HIV-unexposed and HIV-exposed-uninfected infants from 2015 to 2017, were selected for high-throughput next-generation sequencing of the RSV F and G glycoprotein genes. The RSV G and F sequences were analyzed by a bioinformatic pipeline and compared to the USA samples from the same three-year period.
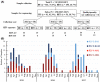
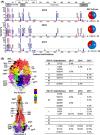
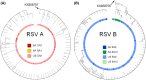
Results: Both RSV A and RSV B co-circulated in South Africa during 2015-2017, with a shift from RSV A (58%-61% in 2015-2016) to RSV B (69%) in 2017. RSV A ON1 and RSV B BA9 genotypes emerged as the most prevalent genotypes in 2017. Variations at the F protein antigenic sites were observed for both RSV A and B strains, with dominant changes (L172Q/S173L) at antigenic site V observed in RSV B strains. RSV A and B F protein sequences from South Africa were very similar to the USA isolates except for a higher rate of RSV A NA1 and RSV B BA10 genotypes in South Africa.
Conclusion: RSV G and F genes continue to evolve and exhibit both local and global circulation patterns in South Africa, supporting the need for continued national surveillance.
Keywords: G and F proteins; RSV; antigenic sites; monoclonal antibodies.
© 2020 The Authors. Influenza and Other Respiratory Viruses Published by John Wiley & Sons Ltd.
Conflict of interest statement
HL, BL, DET, AT, DW, HJ, and MTE are employees and shareholders of AstraZeneca when the study was conducted; SAM, NS and MCN had no competing interests.
Figures

Similar articles
-
Genetic characterization respiratory syncytial virus in Kerala, the southern part of India.J Med Virol. 2017 Dec;89(12):2092-2097. doi: 10.1002/jmv.24842. Epub 2017 Aug 29. J Med Virol. 2017. PMID: 28464224
-
Replacement and positive evolution of subtype A and B respiratory syncytial virus G-protein genotypes from 1997-2012 in South Africa.J Infect Dis. 2013 Dec 15;208 Suppl 3:S227-37. doi: 10.1093/infdis/jit477. J Infect Dis. 2013. PMID: 24265482
-
Global Molecular Epidemiology of Respiratory Syncytial Virus from the 2017-2018 INFORM-RSV Study.J Clin Microbiol. 2020 Dec 17;59(1):e01828-20. doi: 10.1128/JCM.01828-20. Print 2020 Dec 17. J Clin Microbiol. 2020. PMID: 33087438 Free PMC article.
-
Prevalence of human respiratory syncytial virus circulating in Iran.J Infect Public Health. 2016 Mar-Apr;9(2):125-35. doi: 10.1016/j.jiph.2015.05.005. Epub 2015 Jul 2. J Infect Public Health. 2016. PMID: 26143136 Review.
-
Differences Between RSV A and RSV B Subgroups and Implications for Pharmaceutical Preventive Measures.Infect Dis Ther. 2024 Aug;13(8):1725-1742. doi: 10.1007/s40121-024-01012-2. Epub 2024 Jul 6. Infect Dis Ther. 2024. PMID: 38971918 Free PMC article. Review.
Cited by
-
Genomic epidemiology and evolutionary dynamics of respiratory syncytial virus group B in Kilifi, Kenya, 2015-17.Virus Evol. 2020 Jul 15;6(2):veaa050. doi: 10.1093/ve/veaa050. eCollection 2020 Jul. Virus Evol. 2020. PMID: 32913665 Free PMC article.
-
Clinical and biological consequences of respiratory syncytial virus genetic diversity.Ther Adv Infect Dis. 2022 Oct 8;9:20499361221128091. doi: 10.1177/20499361221128091. eCollection 2022 Jan-Dec. Ther Adv Infect Dis. 2022. PMID: 36225856 Free PMC article. Review.
-
Characterisation of RSV Fusion Proteins from South African Patients with RSV Disease, 2019 to 2020.Viruses. 2022 Oct 22;14(11):2321. doi: 10.3390/v14112321. Viruses. 2022. PMID: 36366419 Free PMC article.
-
Molecular Diversity of Human Respiratory Syncytial Virus before and during the COVID-19 Pandemic in Two Neighboring Japanese Cities.Microbiol Spectr. 2023 Aug 17;11(4):e0260622. doi: 10.1128/spectrum.02606-22. Epub 2023 Jul 6. Microbiol Spectr. 2023. PMID: 37409937 Free PMC article.
-
Whole genome molecular analysis of respiratory syncytial virus pre and during the COVID-19 pandemic in Free State province, South Africa.Virus Res. 2024 Sep;347:199421. doi: 10.1016/j.virusres.2024.199421. Epub 2024 Jul 5. Virus Res. 2024. PMID: 38942296 Free PMC article.
References
-
- Collarini EJ, Lee FE, Foord O, et al. Potent high‐affinity antibodies for treatment and prophylaxis of respiratory syncytial virus derived from B cells of infected patients. J Immunol. 2009;183(10):6338‐6345. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

