Epidemiology of β-Lactamase-Producing Pathogens
- PMID: 32102899
- PMCID: PMC7048014
- DOI: 10.1128/CMR.00047-19
Epidemiology of β-Lactamase-Producing Pathogens
Abstract
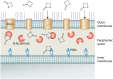
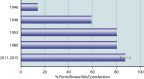
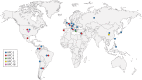
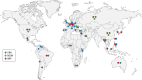
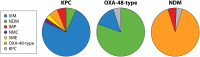
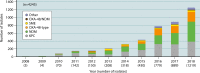
β-Lactam antibiotics have been widely used as therapeutic agents for the past 70 years, resulting in emergence of an abundance of β-lactam-inactivating β-lactamases. Although penicillinases in Staphylococcus aureus challenged the initial uses of penicillin, β-lactamases are most important in Gram-negative bacteria, particularly in enteric and nonfermentative pathogens, where collectively they confer resistance to all β-lactam-containing antibiotics. Critical β-lactamases are those enzymes whose genes are encoded on mobile elements that are transferable among species. Major β-lactamase families include plasmid-mediated extended-spectrum β-lactamases (ESBLs), AmpC cephalosporinases, and carbapenemases now appearing globally, with geographic preferences for specific variants. CTX-M enzymes include the most common ESBLs that are prevalent in all areas of the world. In contrast, KPC serine carbapenemases are present more frequently in the Americas, the Mediterranean countries, and China, whereas NDM metallo-β-lactamases are more prevalent in the Indian subcontinent and Eastern Europe. As selective pressure from β-lactam use continues, multiple β-lactamases per organism are increasingly common, including pathogens carrying three different carbapenemase genes. These organisms may be spread throughout health care facilities as well as in the community, warranting close attention to increased infection control measures and stewardship of the β-lactam-containing drugs in an effort to control selection of even more deleterious pathogens.
Keywords: ESBL; beta-lactamase; carbapenemase; epidemiology; resistance.
Copyright © 2020 American Society for Microbiology.
Figures
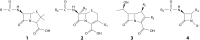
Similar articles
-
Evolution and epidemiology of extended-spectrum beta-lactamases (ESBLs) and ESBL-producing microorganisms.Clin Microbiol Infect. 2001 Nov;7(11):597-608. doi: 10.1046/j.1198-743x.2001.00330.x. Clin Microbiol Infect. 2001. PMID: 11737084 Review.
-
Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae.Curr Opin Microbiol. 2010 Oct;13(5):558-64. doi: 10.1016/j.mib.2010.09.006. Epub 2010 Oct 1. Curr Opin Microbiol. 2010. PMID: 20920882 Review.
-
What's new in antibiotic resistance? Focus on beta-lactamases.Drug Resist Updat. 2006 Jun;9(3):142-56. doi: 10.1016/j.drup.2006.05.005. Epub 2006 Aug 8. Drug Resist Updat. 2006. PMID: 16899402 Review.
-
Molecular epidemiology of extended-spectrum β-lactamase-, AmpC β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007-11.J Antimicrob Chemother. 2013 May;68 Suppl 1:i57-65. doi: 10.1093/jac/dkt027. J Antimicrob Chemother. 2013. PMID: 23587779
-
[MULTIRESISTANT BACTERIA].Acta Med Croatica. 2015 Sep;69(3):211-6. Acta Med Croatica. 2015. PMID: 29077379 Review. Croatian.
Cited by
-
Molecular Epidemiology of Carbapenemases in Enterobacteriales from Humans, Animals, Food and the Environment.Antibiotics (Basel). 2020 Oct 13;9(10):693. doi: 10.3390/antibiotics9100693. Antibiotics (Basel). 2020. PMID: 33066205 Free PMC article. Review.
-
β-Lactamase and Macrolide Resistance Gene Carriage in Escherichia coli Isolates Among Children Discharged From Inpatient Care in Western Kenya: A Cross-sectional Study.Open Forum Infect Dis. 2024 Jun 3;11(6):ofae307. doi: 10.1093/ofid/ofae307. eCollection 2024 Jun. Open Forum Infect Dis. 2024. PMID: 38938894 Free PMC article.
-
Molecular genotyping reveals multiple carbapenemase genes and unique blaOXA-51-like (oxaAb) alleles among clinically isolated Acinetobacter baumannii from a Philippine tertiary hospital.Trop Med Health. 2024 Sep 26;52(1):62. doi: 10.1186/s41182-024-00629-w. Trop Med Health. 2024. PMID: 39327611 Free PMC article.
-
Clinical carbapenem-resistant Klebsiella pneumoniae isolates simultaneously harboring bla NDM-1, bla OXA types and qnrS genes from the Kingdom of Bahrain: Resistance profile and genetic environment.Front Cell Infect Microbiol. 2022 Oct 11;12:1033305. doi: 10.3389/fcimb.2022.1033305. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36304935 Free PMC article.
-
Genomic and Phenotypic Analysis of bla KPC-2 Associated Carbapenem Resistance in Klebsiella aerogenes: Insights into Clonal Spread and Resistance Mechanisms Across Hospital Departments in Beijing.Infect Drug Resist. 2024 Jul 3;17:2735-2749. doi: 10.2147/IDR.S458182. eCollection 2024. Infect Drug Resist. 2024. PMID: 38974314 Free PMC article.
References
-
- ECDC. 2018. Surveillance of antimicrobial resistance in Europe–annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017. European Centre for Disease Prevention and Control, Stockholm, Sweden.
-
- WHO. 2019. No time to wait: securing the future from drug-resistant infections. Report to the Secretary-General of the United Nations. World Health Organization, Geneva, Switzerland.
-
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi:10.1128/AAC.00774-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

