Probing Mechanisms of Transcription Elongation Through Cell-to-Cell Variability of RNA Polymerase
- PMID: 32101716
- PMCID: PMC7136280
- DOI: 10.1016/j.bpj.2020.02.002
Probing Mechanisms of Transcription Elongation Through Cell-to-Cell Variability of RNA Polymerase
Abstract
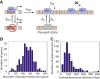
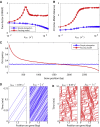
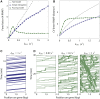
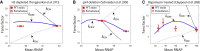
The process of transcription initiation and elongation are primary points of control in the regulation of gene expression. Although biochemical studies have uncovered the mechanisms involved in controlling transcription at each step, how these mechanisms manifest in vivo at the level of individual genes is still unclear. Recent experimental advances have enabled single-cell measurements of RNA polymerase (RNAP) molecules engaged in the process of transcribing a gene of interest. In this article, we use Gillespie simulations to show that measurements of cell-to-cell variability of RNAP numbers and interpolymerase distances can reveal the prevailing mode of regulation of a given gene. Mechanisms of regulation at each step, from initiation to elongation dynamics, produce qualitatively distinct signatures, which can further be used to discern between them. Most intriguingly, depending on the initiation kinetics, stochastic elongation can either enhance or suppress cell-to-cell variability at the RNAP level. To demonstrate the value of this framework, we analyze RNAP number distribution data for ribosomal genes in Saccharomyces cerevisiae from three previously published studies and show that this approach provides crucial mechanistic insights into the transcriptional regulation of these genes.
Copyright © 2020 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Transcription factor dynamics.Microbiology (Reading). 2008 Jul;154(Pt 7):1837-1844. doi: 10.1099/mic.0.2008/018549-0. Microbiology (Reading). 2008. PMID: 18599813 Review.
-
Bacterial RNA polymerase can retain σ70 throughout transcription.Proc Natl Acad Sci U S A. 2016 Jan 19;113(3):602-7. doi: 10.1073/pnas.1513899113. Epub 2016 Jan 5. Proc Natl Acad Sci U S A. 2016. PMID: 26733675 Free PMC article.
-
An Escherichia coli RNA polymerase defective in transcription due to its overproduction of abortive initiation products.J Mol Biol. 1994 Feb 11;236(1):72-80. doi: 10.1006/jmbi.1994.1119. J Mol Biol. 1994. PMID: 7508986
-
Distribution of Initiation Times Reveals Mechanisms of Transcriptional Regulation in Single Cells.Biophys J. 2018 May 8;114(9):2072-2082. doi: 10.1016/j.bpj.2018.03.031. Biophys J. 2018. PMID: 29742401 Free PMC article.
-
RNA polymerase: the vehicle of transcription.Trends Microbiol. 2008 Mar;16(3):126-34. doi: 10.1016/j.tim.2007.12.006. Epub 2008 Feb 14. Trends Microbiol. 2008. PMID: 18280161 Review.
Cited by
-
Real-time single-cell characterization of the eukaryotic transcription cycle reveals correlations between RNA initiation, elongation, and cleavage.PLoS Comput Biol. 2021 May 18;17(5):e1008999. doi: 10.1371/journal.pcbi.1008999. eCollection 2021 May. PLoS Comput Biol. 2021. PMID: 34003867 Free PMC article.
-
Regulation of the dynamic RNA Pol II elongation rate in Drosophila embryos.Cell Rep. 2023 Oct 31;42(10):113225. doi: 10.1016/j.celrep.2023.113225. Epub 2023 Oct 12. Cell Rep. 2023. PMID: 37837623 Free PMC article.
-
Model-based characterization of the equilibrium dynamics of transcription initiation and promoter-proximal pausing in human cells.Nucleic Acids Res. 2023 Nov 27;51(21):e106. doi: 10.1093/nar/gkad843. Nucleic Acids Res. 2023. PMID: 37889042 Free PMC article.
-
Collective polymerase dynamics emerge from DNA supercoiling during transcription.Biophys J. 2022 Nov 1;121(21):4153-4165. doi: 10.1016/j.bpj.2022.09.026. Epub 2022 Sep 27. Biophys J. 2022. PMID: 36171726 Free PMC article.
-
A matter of time: Using dynamics and theory to uncover mechanisms of transcriptional bursting.Curr Opin Cell Biol. 2020 Dec;67:147-157. doi: 10.1016/j.ceb.2020.08.001. Epub 2020 Nov 24. Curr Opin Cell Biol. 2020. PMID: 33242838 Free PMC article. Review.
References
-
- Shandilya J., Roberts S.G.E. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim. Biophys. Acta. 2012;1819:391–400. - PubMed
-
- Svejstrup J.Q. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim. Biophys. Acta. 2004;1677:64–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

