Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2
- PMID: 32100877
- PMCID: PMC7228221
- DOI: 10.1002/jmv.25726
Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2
Abstract
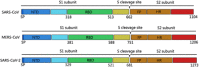
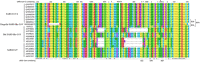
From the beginning of 2002 and 2012, severe respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) crossed the species barriers to infect humans, causing thousands of infections and hundreds of deaths, respectively. Currently, a novel coronavirus (SARS-CoV-2), which has become the cause of the outbreak of Coronavirus Disease 2019 (COVID-19), was discovered. Until 18 February 2020, there were 72 533 confirmed COVID-19 cases (including 10 644 severe cases) and 1872 deaths in China. SARS-CoV-2 is spreading among the public and causing substantial burden due to its human-to-human transmission. However, the intermediate host of SARS-CoV-2 is still unclear. Finding the possible intermediate host of SARS-CoV-2 is imperative to prevent further spread of the epidemic. In this study, we used systematic comparison and analysis to predict the interaction between the receptor-binding domain (RBD) of coronavirus spike protein and the host receptor, angiotensin-converting enzyme 2 (ACE2). The interaction between the key amino acids of S protein RBD and ACE2 indicated that, other than pangolins and snakes, as previously suggested, turtles (Chrysemys picta bellii, Chelonia mydas, and Pelodiscus sinensis) may act as the potential intermediate hosts transmitting SARS-CoV-2 to humans.
Keywords: Coronavirus Disease 2019 (COVID-19); SARS-CoV-2; SARS-CoV-2 spike protein (S); angiotensin-converting enzyme 2 (ACE2); receptor-binding domain (RBD) of coronavirus.
© 2020 Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures


Comment in
-
Coronavirus uses as binding site in humans angiotensin-converting enzyme 2 functional receptor that is involved in arterial blood pressure control and fibrotic response to damage and is a drug target in cardiovascular disease. Is this just a phylogenetic coincidence?J Med Virol. 2020 Oct;92(10):1713-1714. doi: 10.1002/jmv.25774. Epub 2020 Mar 26. J Med Virol. 2020. PMID: 32216172 Free PMC article. No abstract available.
Similar articles
-
Structural basis of receptor recognition by SARS-CoV-2.Nature. 2020 May;581(7807):221-224. doi: 10.1038/s41586-020-2179-y. Epub 2020 Mar 30. Nature. 2020. PMID: 32225175 Free PMC article.
-
Role of the GTNGTKR motif in the N-terminal receptor-binding domain of the SARS-CoV-2 spike protein.Virus Res. 2020 Sep;286:198058. doi: 10.1016/j.virusres.2020.198058. Epub 2020 Jun 9. Virus Res. 2020. PMID: 32531235 Free PMC article.
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Mass Spectrometry and Structural Biology Techniques in the Studies on the Coronavirus-Receptor Interaction.Molecules. 2020 Sep 10;25(18):4133. doi: 10.3390/molecules25184133. Molecules. 2020. PMID: 32927621 Free PMC article. Review.
-
Molecular Basis of Pathogenesis of Coronaviruses: A Comparative Genomics Approach to Planetary Health to Prevent Zoonotic Outbreaks in the 21st Century.OMICS. 2020 Nov;24(11):634-644. doi: 10.1089/omi.2020.0131. Epub 2020 Sep 16. OMICS. 2020. PMID: 32940573 Review.
Cited by
-
A review on Promising vaccine development progress for COVID-19 disease.Vacunas. 2020 Jul-Dec;21(2):121-128. doi: 10.1016/j.vacun.2020.05.002. Epub 2020 Jun 13. Vacunas. 2020. PMID: 32837460 Free PMC article. Review.
-
The 2019-2020 Novel Coronavirus (Severe Acute Respiratory Syndrome Coronavirus 2) Pandemic: A Joint American College of Academic International Medicine-World Academic Council of Emergency Medicine Multidisciplinary COVID-19 Working Group Consensus Paper.J Glob Infect Dis. 2020 May 22;12(2):47-93. doi: 10.4103/jgid.jgid_86_20. eCollection 2020 Apr-Jun. J Glob Infect Dis. 2020. PMID: 32773996 Free PMC article.
-
Analysis of Possible Intermediate Hosts of the New Coronavirus SARS-CoV-2.Front Vet Sci. 2020 Jun 9;7:379. doi: 10.3389/fvets.2020.00379. eCollection 2020. Front Vet Sci. 2020. PMID: 32582786 Free PMC article. No abstract available.
-
Variant analysis of SARS-CoV-2 genomes.Bull World Health Organ. 2020 Jul 1;98(7):495-504. doi: 10.2471/BLT.20.253591. Epub 2020 Jun 2. Bull World Health Organ. 2020. PMID: 32742035 Free PMC article.
-
COVID-19 and New-Onset Psychosis: A Comprehensive Review.J Pers Med. 2023 Jan 2;13(1):104. doi: 10.3390/jpm13010104. J Pers Med. 2023. PMID: 36675765 Free PMC article. Review.
References
-
- Gorbalenya AE, Baker SC, Baric RS, et al. Severe acute respiratory syndrome‐related coronavirus: the species and its viruses—A statement of the Coronavirus Study Group. bioRxiv. 2020. 10.1101/2020.02.07.937862 - DOI
-
- Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276‐278. - PubMed
-
- Drosten C, Kellam P, Memish ZA. Evidence for camel‐to‐human transmission of MERS coronavirus. N Engl J Med. 2014;371(14):1359‐1360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 81471942/National Natural Science Foundation of China/International
- 81730061/National Natural Science Foundation of China/International
- 81902066/National Natural Science Foundation of China/International
- 2018CFB093/Hubei Provincial Natural Science Foundation/International
- 2018CFB185/Hubei Provincial Natural Science Foundation/International
- Q20192104/Natural Science Foundation of Hubei Provincial Department of Education/International
- Q20192105/Natural Science Foundation of Hubei Provincial Department of Education/International
- 2016QDJZR03/Cultivating Project for Young Scholar at Hubei University of Medicine/International
- 2017QDJZR08/Cultivating Project for Young Scholar at Hubei University of Medicine/International
- S201910929032/Hubei Provincial Training Program of Innovation and Entrepreneurship for Undergraduates/International
- S201913249006/Hubei Provincial Training Program of Innovation and Entrepreneurship for Undergraduates/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

