High Levels of Class I Major Histocompatibility Complex mRNA Are Present in Epstein-Barr Virus-Associated Gastric Adenocarcinomas
- PMID: 32098275
- PMCID: PMC7072773
- DOI: 10.3390/cells9020499
High Levels of Class I Major Histocompatibility Complex mRNA Are Present in Epstein-Barr Virus-Associated Gastric Adenocarcinomas
Abstract
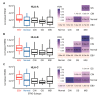
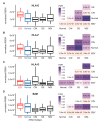
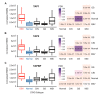
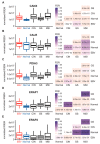
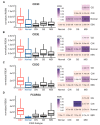
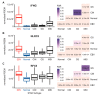
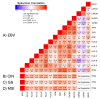
Epstein-Barr virus (EBV) is responsible for approximately 9% of stomach adenocarcinomas. EBV-encoded microRNAs have been reported as reducing the function of the class I major histocompatibility complex (MHC-I) antigen presentation apparatus, which could allow infected cells to evade adaptive immune responses. Using data from nearly 400 human gastric carcinomas (GCs), we assessed the impact of EBV on MHC-I heavy and light chain mRNA levels, as well as multiple other components essential for antigen processing and presentation. Unexpectedly, mRNA levels of these genes were as high, or higher, in EBV-associated gastric carcinomas (EBVaGCs) compared to normal control tissues or other GC subtypes. This coordinated upregulation could have been a consequence of the higher intratumoral levels of interferon γ in EBVaGCs, which correlated with signatures of increased infiltration by T and natural killer (NK) cells. These results indicate that EBV-encoded products do not effectively reduce mRNA levels of the MHC-I antigen presentation apparatus in human GCs.
Keywords: EBV; EBVaGC; Epstein–Barr virus; MHC-I; TCGA; antigen presentation; immune evasion; major histocompatibility complex; stomach adenocarcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
High MHC-II expression in Epstein-Barr virus-associated gastric cancers suggests that tumor cells serve an important role in antigen presentation.Sci Rep. 2020 Sep 8;10(1):14786. doi: 10.1038/s41598-020-71775-4. Sci Rep. 2020. PMID: 32901107 Free PMC article.
-
Down-regulation of locus-specific human lymphocyte antigen class I expression in Epstein-Barr virus-associated gastric cancer: implication for viral-induced immune evasion.Cancer. 2006 Apr 15;106(8):1685-93. doi: 10.1002/cncr.21784. Cancer. 2006. PMID: 16541432
-
Latent Expression of the Epstein-Barr Virus (EBV)-Encoded Major Histocompatibility Complex Class I TAP Inhibitor, BNLF2a, in EBV-Positive Gastric Carcinomas.J Virol. 2015 Oct;89(19):10110-4. doi: 10.1128/JVI.01110-15. Epub 2015 Jul 15. J Virol. 2015. PMID: 26178981 Free PMC article.
-
The tumor immune composition of mismatch repair deficient and Epstein-Barr virus-positive gastric cancer: A systematic review.Cancer Treat Rev. 2024 Jun;127:102737. doi: 10.1016/j.ctrv.2024.102737. Epub 2024 Apr 20. Cancer Treat Rev. 2024. PMID: 38669788 Review.
-
Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review.
Cited by
-
EBV-microRNAs as Potential Biomarkers in EBV-related Fever: A Narrative Review.Curr Mol Med. 2024;24(1):2-13. doi: 10.2174/1566524023666221118122005. Curr Mol Med. 2024. PMID: 36411555 Free PMC article. Review.
-
EBNA2 driven enhancer switching at the CIITA-DEXI locus suppresses HLA class II gene expression during EBV infection of B-lymphocytes.PLoS Pathog. 2021 Aug 5;17(8):e1009834. doi: 10.1371/journal.ppat.1009834. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34352044 Free PMC article.
-
Overview of Epstein-Barr-Virus-Associated Gastric Cancer Correlated with Prognostic Classification and Development of Therapeutic Options.Int J Mol Sci. 2020 Dec 10;21(24):9400. doi: 10.3390/ijms21249400. Int J Mol Sci. 2020. PMID: 33321820 Free PMC article. Review.
-
High MHC-II expression in Epstein-Barr virus-associated gastric cancers suggests that tumor cells serve an important role in antigen presentation.Sci Rep. 2020 Sep 8;10(1):14786. doi: 10.1038/s41598-020-71775-4. Sci Rep. 2020. PMID: 32901107 Free PMC article.
-
The EBV Gastric Cancer Resource (EBV-GCR): A Suite of Tools for Investigating EBV-Associated Human Gastric Carcinogenesis.Viruses. 2023 Mar 27;15(4):853. doi: 10.3390/v15040853. Viruses. 2023. PMID: 37112833 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

