Investigating the Effects of Osmolytes and Environmental pH on Bacterial Persisters
- PMID: 32094133
- PMCID: PMC7179588
- DOI: 10.1128/AAC.02393-19
Investigating the Effects of Osmolytes and Environmental pH on Bacterial Persisters
Abstract
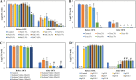
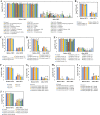
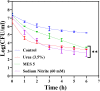
Bacterial persisters are phenotypic variants that temporarily demonstrate an extraordinary tolerance toward antibiotics. Persisters have been linked to the recalcitrance of biofilm-related infections; hence, a complete understanding of their physiology can lead to improvement of therapeutic strategies for such infections. Mechanisms pertaining to persister formation are thought to be associated with stress response pathways triggered by intra- or extracellular stress factors. Unfortunately, studies demonstrating the effects of osmolyte- and/or pH-induced stresses on bacterial persistence are largely missing. To fill this knowledge gap within the field, we studied the effects of various osmolytes and pH conditions on Escherichia coli persistence with the use of phenotype microarrays and antibiotic tolerance assays. Although we found that a number of chemicals and pH environments, including urea, sodium nitrite, and acidic pH, significantly reduced persister formation in E. coli compared to no-osmolyte/no-buffer controls, this reduction in persister levels was less pronounced in late-stationary-phase cultures. Our results further demonstrated a positive correlation between cell growth and persister formation, which challenges the general notion in the field that slow-growing cultures have more persister cells than fast-growing cultures.
Keywords: Escherichia coli; antimicrobial activity; growth rate; osmolytes; pH; persisters; sodium nitrite; tolerance; urea.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
Prophages and Growth Dynamics Confound Experimental Results with Antibiotic-Tolerant Persister Cells.mBio. 2017 Dec 12;8(6):e01964-17. doi: 10.1128/mBio.01964-17. mBio. 2017. PMID: 29233898 Free PMC article.
-
Persister Escherichia coli Cells Have a Lower Intracellular pH than Susceptible Cells but Maintain Their pH in Response to Antibiotic Treatment.mBio. 2021 Aug 31;12(4):e0090921. doi: 10.1128/mBio.00909-21. Epub 2021 Jul 20. mBio. 2021. PMID: 34281389 Free PMC article.
-
Stationary-Phase Persisters to Ofloxacin Sustain DNA Damage and Require Repair Systems Only during Recovery.mBio. 2015 Sep 1;6(5):e00731-15. doi: 10.1128/mBio.00731-15. mBio. 2015. PMID: 26330511 Free PMC article.
-
Multidrug tolerance of biofilms and persister cells.Curr Top Microbiol Immunol. 2008;322:107-31. doi: 10.1007/978-3-540-75418-3_6. Curr Top Microbiol Immunol. 2008. PMID: 18453274 Review.
-
Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics.Drug Resist Updat. 2018 May;38:12-26. doi: 10.1016/j.drup.2018.03.002. Epub 2018 Apr 10. Drug Resist Updat. 2018. PMID: 29857815 Review.
Cited by
-
Cellular Self-Digestion and Persistence in Bacteria.Microorganisms. 2021 Oct 31;9(11):2269. doi: 10.3390/microorganisms9112269. Microorganisms. 2021. PMID: 34835393 Free PMC article. Review.
-
Undecanoic Acid, Lauric Acid, and N-Tridecanoic Acid Inhibit Escherichia coli Persistence and Biofilm Formation.J Microbiol Biotechnol. 2021 Jan 28;31(1):130-136. doi: 10.4014/jmb.2008.08027. J Microbiol Biotechnol. 2021. PMID: 33046677 Free PMC article.
-
Molecular mechanism and application of emerging technologies in study of bacterial persisters.BMC Microbiol. 2024 Nov 16;24(1):480. doi: 10.1186/s12866-024-03628-3. BMC Microbiol. 2024. PMID: 39548389 Free PMC article. Review.
-
Microbiotal characteristics colonized in intestinal mucosa of mice with diarrhoea and repeated stress.3 Biotech. 2020 Aug;10(8):372. doi: 10.1007/s13205-020-02368-1. Epub 2020 Aug 3. 3 Biotech. 2020. PMID: 32832332 Free PMC article.
-
Matrix is everywhere: extracellular DNA is a link between biofilm and mineralization in Bacillus cereus planktonic lifestyle.NPJ Biofilms Microbiomes. 2023 Feb 28;9(1):9. doi: 10.1038/s41522-023-00377-5. NPJ Biofilms Microbiomes. 2023. PMID: 36854956 Free PMC article.
References
-
- Hobby GL, Meyer K, Chaffee E. 1942. Observations on the mechanism of action of penicillin. Exp Biol Med 50:281–285. doi:10.3181/00379727-50-13773. - DOI
-
- Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi:10.1016/S0140-6736(00)74210-3. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

