Type 1 Interferon Responses Underlie Tumor-Selective Replication of Oncolytic Measles Virus
- PMID: 32087150
- PMCID: PMC7132605
- DOI: 10.1016/j.ymthe.2020.01.027
Type 1 Interferon Responses Underlie Tumor-Selective Replication of Oncolytic Measles Virus
Abstract
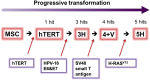
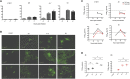
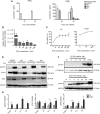
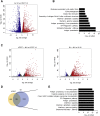
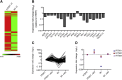
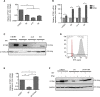
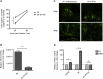
The mechanism of tumor-selective replication of oncolytic measles virus (MV) is poorly understood. Using a stepwise model of cellular transformation, in which oncogenic hits were additively expressed in human bone marrow-derived mesenchymal stromal cells, we show that MV-induced oncolysis increased progressively with transformation. The type 1 interferon (IFN) response to MV infection was significantly reduced and delayed, in accordance with the level of transformation. Consistently, we observed delayed and reduced signal transducer and activator of transcription (STAT1) phosphorylation in the fully transformed cells. Pre-treatment with IFNβ restored resistance to MV-mediated oncolysis. Gene expression profiling to identify the genetic correlates of susceptibility to MV oncolysis revealed a dampened basal level of immune-related genes in the fully transformed cells compared to their normal counterparts. IFN-induced transmembrane protein 1 (IFITM1) was the foremost basally downregulated immune gene. Stable IFITM1 overexpression in MV-susceptible cells resulted in a 50% increase in cell viability and a significant reduction in viral replication at 24 h after MV infection. Overall, our data indicate that the basal reduction in functions of the type 1 IFN pathway is a major contributor to the oncolytic selectivity of MV. In particular, we have identified IFITM1 as a restriction factor for oncolytic MV, acting at early stages of infection.
Keywords: IFITM1; ISG; MV; innate immune response; interferon-induced transmembrane protein 1; interferon-stimulated gene; measles virus; oncolytic measles virus; type 1 interferon.
Copyright © 2020 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling.J Virol. 2014 May;88(9):5152-64. doi: 10.1128/JVI.03851-13. Epub 2014 Feb 26. J Virol. 2014. PMID: 24574393 Free PMC article.
-
Sensitivity of human pleural mesothelioma to oncolytic measles virus depends on defects of the type I interferon response.Oncotarget. 2015 Dec 29;6(42):44892-904. doi: 10.18632/oncotarget.6285. Oncotarget. 2015. PMID: 26539644 Free PMC article.
-
Innate immune defense defines susceptibility of sarcoma cells to measles vaccine virus-based oncolysis.J Virol. 2013 Mar;87(6):3484-501. doi: 10.1128/JVI.02106-12. Epub 2013 Jan 9. J Virol. 2013. PMID: 23302892 Free PMC article.
-
Measles to the Rescue: A Review of Oncolytic Measles Virus.Viruses. 2016 Oct 22;8(10):294. doi: 10.3390/v8100294. Viruses. 2016. PMID: 27782084 Free PMC article. Review.
-
Viral oncolysis - can insights from measles be transferred to canine distemper virus?Viruses. 2014 Jun 11;6(6):2340-75. doi: 10.3390/v6062340. Viruses. 2014. PMID: 24921409 Free PMC article. Review.
Cited by
-
Mesenchymal stromal cells equipped by IFNα empower T cells with potent anti-tumor immunity.Oncogene. 2022 Mar;41(13):1866-1881. doi: 10.1038/s41388-022-02201-4. Epub 2022 Feb 10. Oncogene. 2022. PMID: 35145233 Free PMC article.
-
Identification of hub genes and their novel diagnostic and prognostic significance in pancreatic adenocarcinoma.Cancer Biol Med. 2021 Aug 17;19(7):1029-46. doi: 10.20892/j.issn.2095-3941.2020.0516. Online ahead of print. Cancer Biol Med. 2021. PMID: 34403221 Free PMC article.
-
Simultaneous Expression of Different Therapeutic Genes by Infection with Multiple Oncolytic HSV-1 Vectors.Biomedicines. 2024 Jul 16;12(7):1577. doi: 10.3390/biomedicines12071577. Biomedicines. 2024. PMID: 39062150 Free PMC article.
-
Transcriptome Analysis of Human Glioblastoma Cells Susceptible to Infection with the Leningrad-16 Vaccine Strain of Measles Virus.Viruses. 2022 Nov 2;14(11):2433. doi: 10.3390/v14112433. Viruses. 2022. PMID: 36366531 Free PMC article.
-
Single-cell RNA sequencing uncovers molecular mechanisms of intravenous immunoglobulin plus methylprednisolone in Kawasaki disease: attenuated monocyte-driven inflammation and improved NK cell cytotoxicity.Front Immunol. 2024 Oct 25;15:1455925. doi: 10.3389/fimmu.2024.1455925. eCollection 2024. Front Immunol. 2024. PMID: 39524437 Free PMC article.
References
-
- Dörig R.E., Marcil A., Chopra A., Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

