Dynamic Genome Editing Using In Vivo Synthesized Donor ssDNA in Escherichia coli
- PMID: 32085579
- PMCID: PMC7072734
- DOI: 10.3390/cells9020467
Dynamic Genome Editing Using In Vivo Synthesized Donor ssDNA in Escherichia coli
Abstract
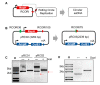
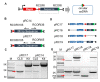
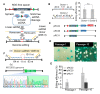
As a key element of genome editing, donor DNA introduces the desired exogenous sequence while working with other crucial machinery such as CRISPR-Cas or recombinases. However, current methods for the delivery of donor DNA into cells are both inefficient and complicated. Here, we developed a new methodology that utilizes rolling circle replication and Cas9 mediated (RC-Cas-mediated) in vivo single strand DNA (ssDNA) synthesis. A single-gene rolling circle DNA replication system from Gram-negative bacteria was engineered to produce circular ssDNA from a Gram-positive parent plasmid at a designed sequence in Escherichia coli. Furthermore, it was demonstrated that the desired linear ssDNA fragment could be cut out using CRISPR-associated protein 9 (CRISPR-Cas9) nuclease and combined with lambda Red recombinase as donor for precise genome engineering. Various donor ssDNA fragments from hundreds to thousands of nucleotides in length were synthesized in E. coli cells, allowing successive genome editing in growing cells. We hope that this RC-Cas-mediated in vivo ssDNA on-site synthesis system will be widely adopted as a useful new tool for dynamic genome editing.
Keywords: PAM-independent; SpyCas9; guide RNA; rolling circle origin; rolling circle replication; single-strand DNA.
Conflict of interest statement
H.Q. is the inventor of one patent application for the biochemical method described in this article. The initial filings were assigned Chinese patent application 201911382846.5. The remaining authors declare no competing financial interests.
Figures

Similar articles
-
Combination of ssDNA recombineering and CRISPR-Cas9 for Pseudomonas putida KT2440 genome editing.Appl Microbiol Biotechnol. 2019 Mar;103(6):2783-2795. doi: 10.1007/s00253-019-09654-w. Epub 2019 Feb 14. Appl Microbiol Biotechnol. 2019. PMID: 30762073
-
Coupling ssDNA recombineering with CRISPR-Cas9 for Escherichia coli DnaG mutations.Appl Microbiol Biotechnol. 2019 Apr;103(8):3559-3570. doi: 10.1007/s00253-019-09744-9. Epub 2019 Mar 16. Appl Microbiol Biotechnol. 2019. PMID: 30879090
-
Metabolic engineering of Escherichia coli BL21 strain using simplified CRISPR-Cas9 and asymmetric homology arms recombineering.Microb Cell Fact. 2022 Feb 5;21(1):19. doi: 10.1186/s12934-022-01746-z. Microb Cell Fact. 2022. PMID: 35123478 Free PMC article.
-
The potential and challenges of CRISPR-Cas in eradication of hepatitis B virus covalently closed circular DNA.Virus Res. 2018 Jan 15;244:304-310. doi: 10.1016/j.virusres.2017.06.010. Epub 2017 Jun 13. Virus Res. 2018. PMID: 28627393 Review.
-
CRISPR-Cas systems: ushering in the new genome editing era.Bioengineered. 2018;9(1):214-221. doi: 10.1080/21655979.2018.1470720. Bioengineered. 2018. PMID: 29968520 Free PMC article. Review.
Cited by
-
Single-stranded circular DNA theranostics.Theranostics. 2022 Jan 1;12(1):35-47. doi: 10.7150/thno.66466. eCollection 2022. Theranostics. 2022. PMID: 34987632 Free PMC article. Review.
References
-
- Codner G.F., Mianné J., Caulder A., Loeffler J., Fell R., King R., Allan A.J., Mackenzie M., Pike F.J., McCabe C.V. Application of long single-stranded DNA donors in genome editing: Generation and validation of mouse mutants. BMC Biol. 2018;16:70. doi: 10.1186/s12915-018-0530-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

