Reciprocal Regulation Between Forkhead Box M1/NF-κB and Methionine Adenosyltransferase 1A Drives Liver Cancer
- PMID: 32080887
- PMCID: PMC7442711
- DOI: 10.1002/hep.31196
Reciprocal Regulation Between Forkhead Box M1/NF-κB and Methionine Adenosyltransferase 1A Drives Liver Cancer
Abstract
Background and aims: Forkhead box M1 (FOXM1) and nuclear factor kappa B (NF-ĸB) are oncogenic drivers in liver cancer that positively regulate each other. We showed that methionine adenosyltransferase 1A (MAT1A) is a tumor suppressor in the liver and inhibits NF-ĸB activity. Here, we examined the interplay between FOXM1/NF-κB and MAT1A in liver cancer.
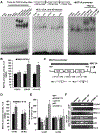
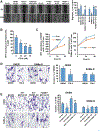
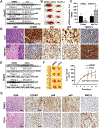
Approach and results: We examined gene and protein expression, effects on promoter activities and binding of proteins to promoter regions, as well as effects of FOXM1 inhibitors T0901317 (T0) and forkhead domain inhibitory-6 (FDI-6) in vitro and in xenograft and syngeneic models of liver cancer. We found, in both hepatocellular carcinoma and cholangiocarcinoma, that an induction in FOXM1 and NF-κB expression is accompanied by a fall in MATα1 (protein encoded by MAT1A). The Cancer Genome Atlas data set confirmed the inverse correlation between FOXM1 and MAT1A. Interestingly, FOXM1 directly interacts with MATα1 and they negatively regulate each other. In contrast, FOXM1 positively regulates p50 and p65 expression through MATα1, given that the effect is lost in its absence. FOXM1, MATα1, and NF-κB all bind to the FOX binding sites in the FOXM1 and MAT1A promoters. However, binding of FOXM1 and NF-κB repressed MAT1A promoter activity, but activated the FOXM1 promoter. In contrast, binding of MATα1 repressed the FOXM1 promoter. MATα1 also binds and represses the NF-κB element in the presence of p65 or p50. Inhibiting FOXM1 with either T0 or FDI-6 inhibited liver cancer cell growth in vitro and in vivo. However, inhibiting FOXM1 had minimal effects in liver cancer cells that do not express MAT1A.
Conclusions: We have found a crosstalk between FOXM1/NF-κB and MAT1A. Up-regulation in FOXM1 lowers MAT1A, but raises NF-κB, expression, and this is a feed-forward loop that enhances tumorigenesis.
© 2020 by the American Association for the Study of Liver Diseases.
Figures





Similar articles
-
Deregulated 14-3-3ζ and methionine adenosyltransferase α1 interplay promotes liver cancer tumorigenesis in mice and humans.Oncogene. 2021 Sep;40(39):5866-5879. doi: 10.1038/s41388-021-01980-6. Epub 2021 Aug 4. Oncogene. 2021. PMID: 34349244 Free PMC article.
-
Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis.J Hepatol. 2013 Oct;59(4):830-41. doi: 10.1016/j.jhep.2013.04.031. Epub 2013 May 7. J Hepatol. 2013. PMID: 23665184 Review.
-
Mechanisms of MAFG Dysregulation in Cholestatic Liver Injury and Development of Liver Cancer.Gastroenterology. 2018 Aug;155(2):557-571.e14. doi: 10.1053/j.gastro.2018.04.032. Epub 2018 May 5. Gastroenterology. 2018. PMID: 29733835 Free PMC article.
-
Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans(‡).Hepatology. 2016 Aug;64(2):439-55. doi: 10.1002/hep.28541. Epub 2016 Apr 28. Hepatology. 2016. PMID: 26969892 Free PMC article.
-
Methionine adenosyltransferases in liver cancer.World J Gastroenterol. 2019 Aug 21;25(31):4300-4319. doi: 10.3748/wjg.v25.i31.4300. World J Gastroenterol. 2019. PMID: 31496615 Free PMC article. Review.
Cited by
-
Comprehensive analysis for cellular senescence-related immunogenic characteristics and immunotherapy prediction of acute myeloid leukemia.Front Pharmacol. 2022 Sep 26;13:987398. doi: 10.3389/fphar.2022.987398. eCollection 2022. Front Pharmacol. 2022. PMID: 36225590 Free PMC article.
-
Hepatic prohibitin 1 and methionine adenosyltransferase α1 defend against primary and secondary liver cancer metastasis.J Hepatol. 2024 Mar;80(3):443-453. doi: 10.1016/j.jhep.2023.11.022. Epub 2023 Dec 10. J Hepatol. 2024. PMID: 38086446
-
Role of transcription factor FOXM1 in diabetes and its complications (Review).Int J Mol Med. 2023 Nov;52(5):101. doi: 10.3892/ijmm.2023.5304. Epub 2023 Sep 8. Int J Mol Med. 2023. PMID: 37681487 Free PMC article.
-
Forkhead Domain Inhibitor-6 Suppresses Corneal Neovascularization and Subsequent Fibrosis After Alkali Burn in Rats.Invest Ophthalmol Vis Sci. 2022 Apr 1;63(4):14. doi: 10.1167/iovs.63.4.14. Invest Ophthalmol Vis Sci. 2022. PMID: 35446346 Free PMC article.
-
MAT1A activation of glycolysis to promote NSCLC progression depends on stabilizing CCND1.Cell Death Dis. 2024 Oct 22;15(10):768. doi: 10.1038/s41419-024-07113-7. Cell Death Dis. 2024. PMID: 39438468 Free PMC article.
References
-
- Wierstra I FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. - PubMed
-
- Yu M, Tang Z, Meng F, Tai M, Zhang J, Wang R, et al. Elevated expression of FoxM1 promotes the tumor cell proliferation in hepatocellular carcinoma. Tumour Biol. 2016;37:1289–97. - PubMed
-
- Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

