Downregulation of S1P Lyase Improves Barrier Function in Human Cerebral Microvascular Endothelial Cells Following an Inflammatory Challenge
- PMID: 32069843
- PMCID: PMC7072972
- DOI: 10.3390/ijms21041240
Downregulation of S1P Lyase Improves Barrier Function in Human Cerebral Microvascular Endothelial Cells Following an Inflammatory Challenge
Abstract
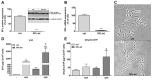
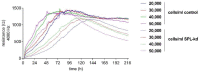
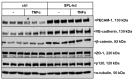
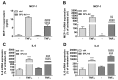
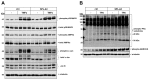
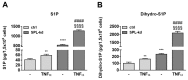
Sphingosine 1-phosphate (S1P) is a key bioactive lipid that regulates a myriad of physiological and pathophysiological processes, including endothelial barrier function, vascular tone, vascular inflammation, and angiogenesis. Various S1P receptor subtypes have been suggested to be involved in the regulation of these processes, whereas the contribution of intracellular S1P (iS1P) through intracellular targets is little explored. In this study, we used the human cerebral microvascular endothelial cell line HCMEC/D3 to stably downregulate the S1P lyase (SPL-kd) and evaluate the consequences on endothelial barrier function and on the molecular factors that regulate barrier tightness under normal and inflammatory conditions. The results show that in SPL-kd cells, transendothelial electrical resistance, as a measure of barrier integrity, was regulated in a dual manner. SPL-kd cells had a delayed barrier build up, a shorter interval of a stable barrier, and, thereafter, a continuous breakdown. Contrariwise, a protection was seen from the rapid proinflammatory cytokine-mediated barrier breakdown. On the molecular level, SPL-kd caused an increased basal protein expression of the adherens junction molecules PECAM-1, VE-cadherin, and β-catenin, increased activity of the signaling kinases protein kinase C, AMP-dependent kinase, and p38-MAPK, but reduced protein expression of the transcription factor c-Jun. However, the only factors that were significantly reduced in TNFα/SPL-kd compared to TNFα/control cells, which could explain the observed protection, were VCAM-1, IL-6, MCP-1, and c-Jun. Furthermore, lipid profiling revealed that dihydro-S1P and S1P were strongly enhanced in TNFα-treated SPL-kd cells. In summary, our data suggest that SPL inhibition is a valid approach to dampenan inflammatory response and augmente barrier integrity during an inflammatory challenge.
Keywords: PKC; S1P lyase; blood–brain barrier; endothelial integrity; inflammation; junctional molecules.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Sphingosine-1-phosphate receptor-2 mediated NFκB activation contributes to tumor necrosis factor-α induced VCAM-1 and ICAM-1 expression in endothelial cells.Prostaglandins Other Lipid Mediat. 2013 Oct;106:62-71. doi: 10.1016/j.prostaglandins.2013.06.001. Epub 2013 Jun 11. Prostaglandins Other Lipid Mediat. 2013. PMID: 23770055 Free PMC article.
-
High-Density Lipoprotein-Associated Apolipoprotein M Limits Endothelial Inflammation by Delivering Sphingosine-1-Phosphate to the Sphingosine-1-Phosphate Receptor 1.Arterioscler Thromb Vasc Biol. 2017 Jan;37(1):118-129. doi: 10.1161/ATVBAHA.116.308435. Epub 2016 Nov 22. Arterioscler Thromb Vasc Biol. 2017. PMID: 27879252
-
Regulation of human cerebro-microvascular endothelial baso-lateral adhesion and barrier function by S1P through dual involvement of S1P1 and S1P2 receptors.Sci Rep. 2016 Jan 27;6:19814. doi: 10.1038/srep19814. Sci Rep. 2016. PMID: 26813587 Free PMC article.
-
Epigenetic regulation of pro-inflammatory cytokine secretion by sphingosine 1-phosphate (S1P) in acute lung injury: Role of S1P lyase.Adv Biol Regul. 2017 Jan;63:156-166. doi: 10.1016/j.jbior.2016.09.007. Epub 2016 Sep 29. Adv Biol Regul. 2017. PMID: 27720306 Free PMC article. Review.
-
S1P Lyase Regulation of Thymic Egress and Oncogenic Inflammatory Signaling.Mediators Inflamm. 2017;2017:7685142. doi: 10.1155/2017/7685142. Epub 2017 Dec 3. Mediators Inflamm. 2017. PMID: 29333002 Free PMC article. Review.
Cited by
-
Regulatory effects of the p38 mitogen-activated protein kinase-myosin light chain kinase pathway on the intestinal epithelial mechanical barrier and the mechanism of modified Pulsatilla decoction in the treatment of ulcerative colitis.J Tradit Chin Med. 2024 Oct;44(5):885-895. doi: 10.19852/j.cnki.jtcm.20240806.006. J Tradit Chin Med. 2024. PMID: 39380219 Free PMC article.
-
Immune System and Brain/Intestinal Barrier Functions in Psychiatric Diseases: Is Sphingosine-1-Phosphate at the Helm?Int J Mol Sci. 2023 Aug 10;24(16):12634. doi: 10.3390/ijms241612634. Int J Mol Sci. 2023. PMID: 37628815 Free PMC article. Review.
-
Lysolipids in Vascular Development, Biology, and Disease.Arterioscler Thromb Vasc Biol. 2021 Feb;41(2):564-584. doi: 10.1161/ATVBAHA.120.305565. Epub 2020 Dec 17. Arterioscler Thromb Vasc Biol. 2021. PMID: 33327749 Free PMC article. Review.
-
Barrier maintenance by S1P during inflammation and sepsis.Tissue Barriers. 2021 Oct 2;9(4):1940069. doi: 10.1080/21688370.2021.1940069. Epub 2021 Jun 21. Tissue Barriers. 2021. PMID: 34152926 Free PMC article. Review.
-
Sphingolipid Metabolism in Glioblastoma and Metastatic Brain Tumors: A Review of Sphingomyelinases and Sphingosine-1-Phosphate.Biomolecules. 2020 Sep 23;10(10):1357. doi: 10.3390/biom10101357. Biomolecules. 2020. PMID: 32977496 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

