Comparative analysis of the viral interferon regulatory factors of KSHV for their requisite for virus production and inhibition of the type I interferon pathway
- PMID: 32056714
- PMCID: PMC7024068
- DOI: 10.1016/j.virol.2019.12.011
Comparative analysis of the viral interferon regulatory factors of KSHV for their requisite for virus production and inhibition of the type I interferon pathway
Abstract
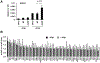
Unique among human viruses, Kaposi's sarcoma-associated herpesvirus (KSHV) encodes several homologs of cellular interferon regulatory factors (vIRFs). Since KSHV expresses multiple factors that can inhibit interferon (IFN) signaling to promote virus production, it is still unclear to what extent vIRFs contribute to these specific processes during KSHV infection. To study the function of vIRFs during viral infection, we engineered 3xFLAG-tagged-vIRF and vIRF-knockout recombinant KSHV clones, which were utilized to test vIRF expression, as well as their requirement for viral replication, virus production, and inhibition of the type I IFN pathway in different models of lytic KSHV infection. Our data show that all vIRFs can be expressed as lytic viral proteins, yet were dispensable for KSHV production and inhibition of type I IFN. Nevertheless, as vIRFs were able to suppress IFN-stimulated antiviral genes, vIRFs may still promote the KSHV lytic cycle in the presence of an ongoing antiviral response.
Keywords: BAC16; IFN signaling; Interferon-stimulated genes; KSHV; Lymphatic endothelial cells; Viral immune evasion; Viral replication; vIRFs.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest There is no conflict of interest.
Figures



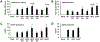
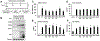
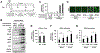
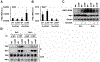
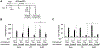
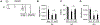
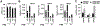
Similar articles
-
Rhesus Macaque Rhadinovirus Encodes a Viral Interferon Regulatory Factor To Disrupt Promyelocytic Leukemia Nuclear Bodies and Antagonize Type I Interferon Signaling.J Virol. 2019 Mar 5;93(6):e02147-18. doi: 10.1128/JVI.02147-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626678 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus vIRF2 protein utilizes an IFN-dependent pathway to regulate viral early gene expression.PLoS Pathog. 2019 May 6;15(5):e1007743. doi: 10.1371/journal.ppat.1007743. eCollection 2019 May. PLoS Pathog. 2019. PMID: 31059555 Free PMC article.
-
A Rhesus Rhadinovirus Viral Interferon (IFN) Regulatory Factor Is Virion Associated and Inhibits the Early IFN Antiviral Response.J Virol. 2015 Aug;89(15):7707-21. doi: 10.1128/JVI.01175-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972548 Free PMC article.
-
Beyond Viral Interferon Regulatory Factors: Immune Evasion Strategies.J Microbiol Biotechnol. 2019 Dec 28;29(12):1873-1881. doi: 10.4014/jmb.1910.10004. J Microbiol Biotechnol. 2019. PMID: 31650769 Review.
-
Viral interferon regulatory factors.J Interferon Cytokine Res. 2009 Sep;29(9):621-7. doi: 10.1089/jir.2009.0067. J Interferon Cytokine Res. 2009. PMID: 19715458 Free PMC article. Review.
Cited by
-
Genome-Wide Transcriptional Roles of KSHV Viral Interferon Regulatory Factors in Oral Epithelial Cells.Viruses. 2024 May 25;16(6):846. doi: 10.3390/v16060846. Viruses. 2024. PMID: 38932139 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Immediate Early Proteins Trigger FOXQ1 Expression in Oral Epithelial Cells, Engaging in a Novel Lytic Cycle-Sustaining Positive Feedback Loop.J Virol. 2023 Mar 30;97(3):e0169622. doi: 10.1128/jvi.01696-22. Epub 2023 Feb 23. J Virol. 2023. PMID: 36815831 Free PMC article.
-
Innate Immune Responses to Herpesvirus Infection.Cells. 2021 Aug 18;10(8):2122. doi: 10.3390/cells10082122. Cells. 2021. PMID: 34440891 Free PMC article. Review.
-
Co-Infection of the Epstein-Barr Virus and the Kaposi Sarcoma-Associated Herpesvirus.Viruses. 2022 Dec 2;14(12):2709. doi: 10.3390/v14122709. Viruses. 2022. PMID: 36560713 Free PMC article. Review.
-
Endoplasmic Reticulum-Shaping Atlastin Proteins Facilitate KSHV Replication.Front Cell Infect Microbiol. 2022 Jan 13;11:790243. doi: 10.3389/fcimb.2021.790243. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35096644 Free PMC article.
References
-
- Aresté C, Blackbourn DJ, 2009. Modulation of the immune system by Kaposi’s sarcoma-associated herpesvirus. Trends Microbiol 17, 119–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

