Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma
- PMID: 32046766
- PMCID: PMC7011553
- DOI: 10.1186/s12967-020-02255-6
Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma
Abstract
Background: Growing evidence has suggested that immune-related genes play crucial roles in the development and progression of hepatocellular carcinoma (HCC). Nevertheless, the utility of immune-related genes for evaluating the prognosis of HCC patients are still lacking. The study aimed to explore gene signatures and prognostic values of immune-related genes in HCC.
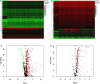
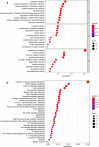
Methods: We comprehensively integrated gene expression data acquired from 374 HCC and 50 normal tissues in The Cancer Genome Atlas (TCGA). Differentially expressed genes (DEGs) analysis and univariate Cox regression analysis were performed to identify DEGs that related to overall survival. An immune prognostic model was constructed using the Lasso and multivariate Cox regression analyses. Furthermore, Cox regression analysis was applied to identify independent prognostic factors in HCC. The correlation analysis between immune-related signature and immune cells infiltration were also investigated. Finally, the signature was validated in an external independent dataset.
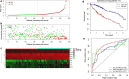
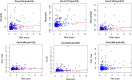
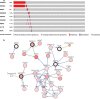
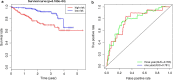
Results: A total of 329 differentially expressed immune-related genes were detected. 64 immune-related genes were identified to be markedly related to overall survival in HCC patients using univariate Cox regression analysis. Then we established a TF-mediated network for exploring the regulatory mechanisms of these genes. Lasso and multivariate Cox regression analyses were applied to construct the immune-based prognostic model, which consisted of nine immune-related genes. Further analysis indicated that this immune-related prognostic model could be an independent prognostic indicator after adjusting to other clinical factors. The relationships between the risk score model and immune cell infiltration suggested that the nine-gene signature could reflect the status of tumor immune microenvironment. The prognostic value of this nine-gene prognostic model was further successfully validated in an independent database.
Conclusions: Together, our study screened potential prognostic immune-related genes and established a novel immune-based prognostic model of HCC, which not only provides new potential prognostic biomarkers and therapeutic targets, but also deepens our understanding of tumor immune microenvironment status and lays a theoretical foundation for immunotherapy.
Keywords: Bioinformatics; Hepatocellular carcinoma; Immune related gene; Prognosis; Prognostic signature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Bioinformatics Analysis of Prognostic Tumor Microenvironment-Related Genes in the Tumor Microenvironment of Hepatocellular Carcinoma.Med Sci Monit. 2020 Mar 31;26:e922159. doi: 10.12659/MSM.922159. Med Sci Monit. 2020. PMID: 32231177 Free PMC article.
-
Identification and Validation of a Novel Six-Gene Expression Signature for Predicting Hepatocellular Carcinoma Prognosis.Front Immunol. 2021 Dec 1;12:723271. doi: 10.3389/fimmu.2021.723271. eCollection 2021. Front Immunol. 2021. PMID: 34925311 Free PMC article.
-
Identification of Prognostic Genes in the Tumor Microenvironment of Hepatocellular Carcinoma.Front Immunol. 2021 Apr 7;12:653836. doi: 10.3389/fimmu.2021.653836. eCollection 2021. Front Immunol. 2021. PMID: 33897701 Free PMC article.
-
Mining Prognostic Biomarkers of Hepatocellular Carcinoma Based on Immune-Associated Genes.DNA Cell Biol. 2020 Apr;39(4):499-512. doi: 10.1089/dna.2019.5099. Epub 2020 Feb 18. DNA Cell Biol. 2020. PMID: 32069130 Review.
-
Identification and validation of immune and prognosis-related genes in hepatocellular carcinoma: A review.Medicine (Baltimore). 2022 Nov 18;101(46):e31814. doi: 10.1097/MD.0000000000031814. Medicine (Baltimore). 2022. PMID: 36401409 Free PMC article. Review.
Cited by
-
Development of a poor-prognostic-mutations derived immune prognostic model for acute myeloid leukemia.Sci Rep. 2021 Mar 1;11(1):4856. doi: 10.1038/s41598-021-84190-0. Sci Rep. 2021. PMID: 33649342 Free PMC article.
-
Development and Validation of a Novel Immune-Gene Pairs Prognostic Model Associated with CTNNB1 Alteration in Hepatocellular Carcinoma.Med Sci Monit. 2020 Sep 18;26:e925494. doi: 10.12659/MSM.925494. Med Sci Monit. 2020. PMID: 32945289 Free PMC article.
-
Construction of a Prognostic Model for Hepatocellular Carcinoma Based on Immunoautophagy-Related Genes and Tumor Microenvironment.Int J Gen Med. 2021 Sep 8;14:5461-5473. doi: 10.2147/IJGM.S325884. eCollection 2021. Int J Gen Med. 2021. PMID: 34526813 Free PMC article.
-
Preliminary Analysis of Aging-Related Genes in Intracerebral Hemorrhage by Integration of Bulk and Single-Cell RNA Sequencing Technology.Int J Gen Med. 2024 Jun 12;17:2719-2740. doi: 10.2147/IJGM.S457480. eCollection 2024. Int J Gen Med. 2024. PMID: 38883702 Free PMC article.
-
Xanthine dehydrogenase as a prognostic biomarker related to tumor immunology in hepatocellular carcinoma.Cancer Cell Int. 2021 Sep 8;21(1):475. doi: 10.1186/s12935-021-02173-7. Cancer Cell Int. 2021. PMID: 34496841 Free PMC article.
References
-
- Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

