Mathematical Properties of Linkage Disequilibrium Statistics Defined by Normalization of the Coefficient D = pAB - pApB
- PMID: 32045910
- PMCID: PMC7199518
- DOI: 10.1159/000504171
Mathematical Properties of Linkage Disequilibrium Statistics Defined by Normalization of the Coefficient D = pAB - pApB
Abstract
Background: Many statistics for measuring linkage disequilibrium (LD) take the form of a normalization of the LD coefficient D. Different normalizations produce statistics with different ranges, interpretations, and arguments favoring their use.
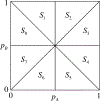
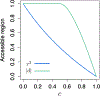
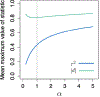
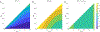
Methods: Here, to compare the mathematical properties of these normalizations, we consider 5 of these normalized statistics, describing their upper bounds, the mean values of their maxima over the set of possible allele frequency pairs, and the size of the allele frequency regions accessible given specified values of the statistics.
Results: We produce detailed characterizations of these properties for the statistics d and ρ, analogous to computations previously performed for r2. We examine the relationships among the statistics, uncovering conditions under which some of them have close connections.
Conclusion: The results contribute insight into LD measurement, particularly the understanding of differences in the features of different LD measures when computed on the same data.
Keywords: Allele frequencies; Linkage disequilibrium; Population genetics; Statistical genetics; Statistics.
© 2020 S. Karger AG, Basel.
Conflict of interest statement
S
The authors have no ethical conflicts to disclose.
D
The authors have no conflicts of interest to declare.
Figures



Similar articles
-
Bounds and normalization of the composite linkage disequilibrium coefficient.Genet Epidemiol. 2004 Nov;27(3):252-7. doi: 10.1002/gepi.20015. Genet Epidemiol. 2004. PMID: 15389931
-
Mathematical properties of the r2 measure of linkage disequilibrium.Theor Popul Biol. 2008 Aug;74(1):130-7. doi: 10.1016/j.tpb.2008.05.006. Epub 2008 Jun 1. Theor Popul Biol. 2008. PMID: 18572214 Free PMC article.
-
The nonlinear structure of linkage disequilibrium.Theor Popul Biol. 2020 Aug;134:160-170. doi: 10.1016/j.tpb.2020.02.005. Epub 2020 Mar 25. Theor Popul Biol. 2020. PMID: 32222435
-
Linkage disequilibrium for different scales and applications.Brief Bioinform. 2004 Dec;5(4):355-64. doi: 10.1093/bib/5.4.355. Brief Bioinform. 2004. PMID: 15606972 Review.
-
On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
Cited by
-
Linkage disequilibrium between rare mutations.Genetics. 2022 Apr 4;220(4):iyac004. doi: 10.1093/genetics/iyac004. Genetics. 2022. PMID: 35100407 Free PMC article.
-
Genome-Wide SNP and Indel Discovery in Abaca (Musa textilis Née) and among Other Musa spp. for Abaca Genetic Resources Management.Curr Issues Mol Biol. 2023 Jul 12;45(7):5776-5797. doi: 10.3390/cimb45070365. Curr Issues Mol Biol. 2023. PMID: 37504281 Free PMC article.
-
Intragenomic conflicts with plasmids and chromosomal mobile genetic elements drive the evolution of natural transformation within species.PLoS Biol. 2024 Oct 14;22(10):e3002814. doi: 10.1371/journal.pbio.3002814. eCollection 2024 Oct. PLoS Biol. 2024. PMID: 39401218 Free PMC article.
-
Single Nucleotide Polymorphisms of CYP3A4 and CYP3A5 in Romanian Kidney Transplant Recipients: Effect on Tacrolimus Pharmacokinetics in a Single-Center Experience.J Clin Med. 2024 Mar 28;13(7):1968. doi: 10.3390/jcm13071968. J Clin Med. 2024. PMID: 38610733 Free PMC article.
-
Genetics of SLE: does this explain susceptibility and severity across racial groups?Rheumatology (Oxford). 2023 Mar 29;62(Suppl 1):i15-i21. doi: 10.1093/rheumatology/keac695. Rheumatology (Oxford). 2023. PMID: 36583554 Free PMC article. Review.
References
-
- Hudson RR: Linkage disequilibrium and recombination; in Balding DJ, Bishop M, Cannings C (eds): Handbook of Statistical Genetics. Chichester, Wiley, 2001, pp 309–324.
-
- Nordborg M, Tavaré S: Linkage disequilibrium: what history has to tell us. Trends Genet 2002; 18: 83–90. - PubMed
-
- McVean G: Linkage disequilibrium, recombination and selection; in Balding DJ, Bishop M, Cannings C (eds): Handbook of Statistical Genetics, ed 3 Chichester, Wiley, 2007, pp 909–944.
-
- Weir BS: Linkage disequilibrium and association mapping. Annu Rev Genomics Hum Genet 2008; 9: 129–142. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

