Integrating transposable elements in the 3D genome
- PMID: 32042316
- PMCID: PMC7001275
- DOI: 10.1186/s13100-020-0202-3
Integrating transposable elements in the 3D genome
Abstract
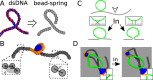
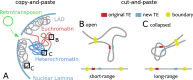
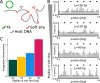
Chromosome organisation is increasingly recognised as an essential component of genome regulation, cell fate and cell health. Within the realm of transposable elements (TEs) however, the spatial information of how genomes are folded is still only rarely integrated in experimental studies or accounted for in modelling. Whilst polymer physics is recognised as an important tool to understand the mechanisms of genome folding, in this commentary we discuss its potential applicability to aspects of TE biology. Based on recent works on the relationship between genome organisation and TE integration, we argue that existing polymer models may be extended to create a predictive framework for the study of TE integration patterns. We suggest that these models may offer orthogonal and generic insights into the integration profiles (or "topography") of TEs across organisms. In addition, we provide simple polymer physics arguments and preliminary molecular dynamics simulations of TEs inserting into heterogeneously flexible polymers. By considering this simple model, we show how polymer folding and local flexibility may generically affect TE integration patterns. The preliminary discussion reported in this commentary is aimed to lay the foundations for a large-scale analysis of TE integration dynamics and topography as a function of the three-dimensional host genome.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures
Similar articles
-
Modeling transposable element dynamics with fragmentation equations.Math Biosci. 2018 Aug;302:46-66. doi: 10.1016/j.mbs.2018.05.009. Epub 2018 May 19. Math Biosci. 2018. PMID: 29787745
-
Contribution of transposable elements in the plant's genome.Gene. 2018 Jul 30;665:155-166. doi: 10.1016/j.gene.2018.04.050. Epub 2018 Apr 22. Gene. 2018. PMID: 29684486 Review.
-
Characterization and functional annotation of nested transposable elements in eukaryotic genomes.Genomics. 2012 Oct;100(4):222-30. doi: 10.1016/j.ygeno.2012.07.004. Epub 2012 Jul 16. Genomics. 2012. PMID: 22800764
-
McClintock: An Integrated Pipeline for Detecting Transposable Element Insertions in Whole-Genome Shotgun Sequencing Data.G3 (Bethesda). 2017 Aug 7;7(8):2763-2778. doi: 10.1534/g3.117.043893. G3 (Bethesda). 2017. PMID: 28637810 Free PMC article.
-
Silencing of Transposable Elements by piRNAs in Drosophila: An Evolutionary Perspective.Genomics Proteomics Bioinformatics. 2017 Jun;15(3):164-176. doi: 10.1016/j.gpb.2017.01.006. Epub 2017 Jun 8. Genomics Proteomics Bioinformatics. 2017. PMID: 28602845 Free PMC article. Review.
Cited by
-
Where the Wild Things Are: Transposable Elements as Drivers of Structural and Functional Variations in the Wheat Genome.Front Plant Sci. 2020 Sep 18;11:585515. doi: 10.3389/fpls.2020.585515. eCollection 2020. Front Plant Sci. 2020. PMID: 33072155 Free PMC article. Review.
-
Plant biosynthetic gene clusters in the context of metabolic evolution.Nat Prod Rep. 2022 Jul 20;39(7):1465-1482. doi: 10.1039/d2np00005a. Nat Prod Rep. 2022. PMID: 35441651 Free PMC article. Review.
-
Transposable Elements: Major Players in Shaping Genomic and Evolutionary Patterns.Cells. 2022 Mar 19;11(6):1048. doi: 10.3390/cells11061048. Cells. 2022. PMID: 35326499 Free PMC article. Review.
-
Investigating site-selection mechanisms of retroviral integration in supercoiled DNA braids.J R Soc Interface. 2021 Aug;18(181):20210229. doi: 10.1098/rsif.2021.0229. Epub 2021 Aug 25. J R Soc Interface. 2021. PMID: 34428944 Free PMC article.
-
Epigenetic conflict on a degenerating Y chromosome increases mutational burden in Drosophila males.Nat Commun. 2020 Nov 2;11(1):5537. doi: 10.1038/s41467-020-19134-9. Nat Commun. 2020. PMID: 33139741 Free PMC article.
References
-
- Schnable PS, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD. Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S. The B73 Maize Genome: Complexity, Diversity, and Dynamics. J Sci. 2009;326:1112. - PubMed
-
- Lee HE, Ayarpadikannan S, Kim HS. Role of transposable elements in genomic rearrangement, evolution, gene regulation and epigenetics in primates. Genes Genet Syst. 2016;90(18805779):245. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

