Genomic Characterization and Phylogenetic Classification of Bovine Coronaviruses Through Whole Genome Sequence Analysis
- PMID: 32041103
- PMCID: PMC7077292
- DOI: 10.3390/v12020183
Genomic Characterization and Phylogenetic Classification of Bovine Coronaviruses Through Whole Genome Sequence Analysis
Abstract
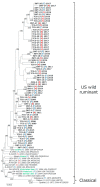
Bovine coronavirus (BCoV) is zoonotically transmissible among species, since BCoV-like viruses have been detected in wild ruminants and humans. BCoV causing enteric and respiratory disease is widespread in cattle farms worldwide; however, limited information is available regarding the molecular characterization of BCoV because of its large genome size, despite its significant economic impact. This study aimed to better understand the genomic characterization and evolutionary dynamics of BCoV via comparative sequence and phylogenetic analyses through whole genome sequence analysis using 67 BCoV isolates collected throughout Japan from 2006 to 2017. On comparing the genomic sequences of the 67 BCoVs, genetic variations were detected in 5 of 10 open reading frames (ORFs) in the BCoV genome. Phylogenetic analysis using whole genomes from the 67 Japanese BCoV isolates in addition to those from 16 reference BCoV strains, revealed the existence of two major genotypes (classical and US wild ruminant genotypes). All Japanese BCoV isolates originated from the US wild ruminant genotype, and they tended to form the same clusters based on the year and farm of collection, not the disease type. Phylogenetic trees on hemagglutinin-esterase protein (HE), spike glycoprotein (S), nucleocapsid protein (N) genes and ORF1 revealed clusters similar to that on whole genome, suggesting that the evolution of BCoVs may be closely associated with variations in these genes. Furthermore, phylogenetic analysis of BCoV S genes including those of European and Asian BCoVs and human enteric coronavirus along with the Japanese BCoVs revealed that BCoVs differentiated into two major types (European and American types). Moreover, the European and American types were divided into eleven and three genotypes, respectively. Our analysis also demonstrated that BCoVs with different genotypes periodically emerged and predominantly circulated within the country. These findings provide useful information to elucidate the detailed molecular characterization of BCoVs, which have spread worldwide. Further genomic analyses of BCoV are essential to deepen the understanding of the evolution of this virus.
Keywords: bovine coronavirus; genotype classification; phylogenetic analysis; spike protein; whole genome.
Conflict of interest statement
We have no conflict of interest to declare.
Figures

Similar articles
-
Detection and molecular characterization of calf diarrhoea bovine coronaviruses circulating in South Korea during 2004-2005.Zoonoses Public Health. 2007;54(6-7):223-30. doi: 10.1111/j.1863-2378.2007.01045.x. Zoonoses Public Health. 2007. PMID: 17803510
-
Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains.J Gen Virol. 2013 Sep;94(Pt 9):2036-2049. doi: 10.1099/vir.0.054940-0. Epub 2013 Jun 26. J Gen Virol. 2013. PMID: 23804565
-
Isolation and Characterization of Contemporary Bovine Coronavirus Strains.Viruses. 2024 Jun 16;16(6):965. doi: 10.3390/v16060965. Viruses. 2024. PMID: 38932257 Free PMC article.
-
Bovine respiratory coronavirus.Vet Clin North Am Food Anim Pract. 2010 Jul;26(2):349-64. doi: 10.1016/j.cvfa.2010.04.005. Vet Clin North Am Food Anim Pract. 2010. PMID: 20619189 Free PMC article. Review.
-
Bovine-like coronaviruses in domestic and wild ruminants.Anim Health Res Rev. 2018 Dec;19(2):113-124. doi: 10.1017/S1466252318000117. Anim Health Res Rev. 2018. PMID: 30683171 Free PMC article. Review.
Cited by
-
A comprehensive molecular analysis of bovine coronavirus strains isolated from Brazil and comparison of a wild-type and cell culture-adapted strain associated with respiratory disease.Braz J Microbiol. 2024 Jun;55(2):1967-1977. doi: 10.1007/s42770-024-01287-0. Epub 2024 Feb 21. Braz J Microbiol. 2024. PMID: 38381350
-
Molecular Detection of Southern Tomato Amalgavirus Prevalent in Tomatoes and Its Genomic Characterization with Global Evolutionary Dynamics.Viruses. 2022 Nov 9;14(11):2481. doi: 10.3390/v14112481. Viruses. 2022. PMID: 36366579 Free PMC article.
-
Bovine Coronavirus and the Associated Diseases.Front Vet Sci. 2021 Mar 31;8:643220. doi: 10.3389/fvets.2021.643220. eCollection 2021. Front Vet Sci. 2021. PMID: 33869323 Free PMC article. Review.
-
SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus.Travel Med Infect Dis. 2020 Sep-Oct;37:101830. doi: 10.1016/j.tmaid.2020.101830. Epub 2020 Aug 2. Travel Med Infect Dis. 2020. PMID: 32755673 Free PMC article. Review.
-
First report and genomic characterization of a bovine-like coronavirus causing enteric infection in an odd-toed non-ruminant species (Indonesian tapir, Acrocodia indica) during an outbreak of winter dysentery in a zoo.Transbound Emerg Dis. 2022 Sep;69(5):3056-3065. doi: 10.1111/tbed.14300. Epub 2021 Aug 31. Transbound Emerg Dis. 2022. PMID: 34427399 Free PMC article.
References
-
- Mebus C.A., Stair E.L., Rhodes M.B., Twiehaus M.J. Neonatal calf diarrhea; propagation, attenuation, and characteristics of coronavirus-like agent. Am. J. Vet. Res. 1973;34:145–150. - PubMed
-
- Storz J., Purdy W., Lin X., Burrell M., Truax R.E., Briggs R.E., Frank G.H., Loan R.W. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp. from cattle involved in two natural outbreaks of shipping fever. J. Am. Vet. Med. Assoc. 2000;216:1599–1604. doi: 10.2460/javma.2000.216.1599. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

