Two Arabidopsis Chloroplast GrpE Homologues Exhibit Distinct Biological Activities and Can Form Homo- and Hetero-Oligomers
- PMID: 32038688
- PMCID: PMC6987454
- DOI: 10.3389/fpls.2019.01719
Two Arabidopsis Chloroplast GrpE Homologues Exhibit Distinct Biological Activities and Can Form Homo- and Hetero-Oligomers
Abstract
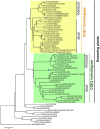
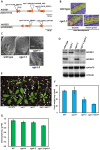
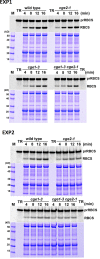
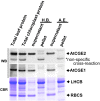
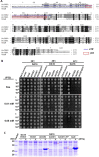
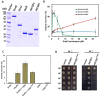
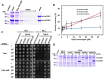
Flowering plants have evolved two distinct clades of chloroplast GrpE homologues (CGEs), which are the nucleotide exchange factor for Hsp70. In Arabidopsis, they are named AtCGE1 (At5g17710) and AtCGE2 (At1g36390). Characterization of their corresponding T-DNA insertion mutants revealed that there is no visible change in phenotype except a defect in protein import in an AtCGE2-knockout mutant under normal growth conditions. However, the embryo development of an AtCGE1-knockout mutant was arrested early at the globular stage. An AtCGE1-knockdown mutant, harboring a T-DNA insertion in the 5'-UTR region, exhibited growth retardation and protein import defect, and its mutant phenotypes became more severe when AtCGE2 was further knocked out. Sub-organellar distribution implied that AtCGE2 might be important for membrane biology due to its preferential association with chloroplast membranes. Biochemical studies and complementation tests showed that only AtCGE1, but not AtCGE2, can effectively rescue the heat-sensitive phenotype of Escherichia coli grpE mutant and robustly stimulate the refolding of denatured luciferase by DnaK. Interestingly, AtCGE1 and AtCGE2 are tending to form heterocomplexes, which exhibit comparable co-chaperone activity to AtCGE1 homocomplexes. Our data indicate that AtCGE1 is the principle functional homologue of GrpE. The possibility that AtCGE2 has a subsidiary or regulatory function through homo- and/or hetero-oligomerization is discussed.
Keywords: DnaK; Hsp70; chloroplast GrpE homologue (CGE); embryo lethal; luciferase refolding assay; oligomerization.
Copyright © 2020 Su, Lin and Lai.
Figures

Similar articles
-
Functional analysis of the Chloroplast GrpE (CGE) proteins from Arabidopsis thaliana.Plant Physiol Biochem. 2019 Jun;139:293-306. doi: 10.1016/j.plaphy.2019.03.027. Epub 2019 Mar 20. Plant Physiol Biochem. 2019. PMID: 30927692
-
Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE.J Mol Biol. 2001 Feb 2;305(5):1173-83. doi: 10.1006/jmbi.2000.4373. J Mol Biol. 2001. PMID: 11162122
-
The heat-sensitive Escherichia coli grpE280 phenotype: impaired interaction of GrpE(G122D) with DnaK.J Mol Biol. 2005 Nov 4;353(4):888-96. doi: 10.1016/j.jmb.2005.08.069. Epub 2005 Sep 20. J Mol Biol. 2005. PMID: 16198374
-
GrpE, a nucleotide exchange factor for DnaK.Cell Stress Chaperones. 2003 Fall;8(3):218-24. doi: 10.1379/1466-1268(2003)008<0218:ganeff>2.0.co;2. Cell Stress Chaperones. 2003. PMID: 14984054 Free PMC article. Review.
-
The Escherichia coli chaperones involved in DNA replication.Philos Trans R Soc Lond B Biol Sci. 1993 Mar 29;339(1289):271-7; discussion 277-8. doi: 10.1098/rstb.1993.0025. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8098531 Review.
Cited by
-
Worldwide Selection Footprints for Drought and Heat in Bread Wheat (Triticum aestivum L.).Plants (Basel). 2022 Sep 1;11(17):2289. doi: 10.3390/plants11172289. Plants (Basel). 2022. PMID: 36079671 Free PMC article.
-
Dipterocarpoidae genomics reveal their demography and adaptations to Asian rainforests.Nat Commun. 2024 Feb 23;15(1):1683. doi: 10.1038/s41467-024-45836-5. Nat Commun. 2024. PMID: 38395938 Free PMC article.
References
-
- Ang D., Chandrasekhar G. N., Zylicz M., Georgopoulos C. (1986). Escherichia coli grpE gene codes for heat shock protein B25.3, essential for both lambda DNA replication at all temperatures and host growth at high temperature. J. Bacteriol. 167 (1), 25–29. 10.1128/jb.167.1.25-29.1986 - DOI - PMC - PubMed
-
- Belmonte M. F., Kirkbride R. C., Stone S. L., Pelletier J. M., Bui A. Q., Yeung E. C., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. U.S.A. 110 (5), E435–E444. 10.1073/pnas.1222061110 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

