Conserved Autophagy Pathway Contributes to Stress Tolerance and Virulence and Differentially Controls Autophagic Flux Upon Nutrient Starvation in Cryptococcus neoformans
- PMID: 32038502
- PMCID: PMC6988817
- DOI: 10.3389/fmicb.2019.02690
Conserved Autophagy Pathway Contributes to Stress Tolerance and Virulence and Differentially Controls Autophagic Flux Upon Nutrient Starvation in Cryptococcus neoformans
Abstract
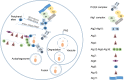
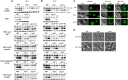
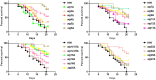
Autophagy is mainly a catabolic process, which is used to cope with nutrient deficiency and various stress conditions. Human environment often imposes various stresses on Cryptococcus neoformans, a major fungal pathogen of immunocompromised individuals; therefore, autophagic response of C. neoformans to these stresses often determines its survival in the host. However, a systematic study on how autophagy related (ATG) genes influence on autophagic flux, virulence, stress response and pathogenicity of C. neoformans is lacking. In this study, 22 ATG-deficient strains were constructed to investigate their roles in virulence, pathogenesis, stress response, starvation tolerance and autophagic flux in C. neoformans. Our results showed that Atg6 and Atg14-03 significantly affect the growth of C. neoformans at 37°C and laccase production. Additionally, atg2Δ and atg6Δ strains were sensitive to oxidative stress caused by hydrogen peroxide. Approximately half of the atgΔ strains displayed higher sensitivity to 1.5 M NaCl and remarkably lower virulence in the Galleria mellonella model than the wild type. Autophagic flux in C. neoformans was dependent on the Atg1-Atg13, Atg5-Atg12-Atg16, and Atg2-Atg18 complexes and Atg11. Cleavage of the green fluorescent protein (GFP) from Atg8 was difficult to detect in these autophagy defective mutants; however, it was detected in the atg3Δ, atg4Δ, atg6Δ and atg14Δ strains. Additionally, no homologs of Saccharomyces cerevisiae ATG10 were detected in C. neoformans. Our results indicate that these ATG genes contribute differentially to carbon and nitrogen starvation tolerance in C. neoformans compared with S. cerevisiae. Overall, this study advances our knowledge of the specific roles of ATG genes in C. neoformans.
Keywords: ATG genes; Atg8; Cryptococcus neoformans; autophagy; starvation; stress tolerance; virulence.
Copyright © 2019 Zhao, Feng, Zhu, Li, Ma, Li, Zhu and Wei.
Figures



Similar articles
-
ATG Genes Influence the Virulence of Cryptococcus neoformans through Contributions beyond Core Autophagy Functions.Infect Immun. 2018 Aug 22;86(9):e00069-18. doi: 10.1128/IAI.00069-18. Print 2018 Sep. Infect Immun. 2018. PMID: 29986893 Free PMC article.
-
PI3K signaling of autophagy is required for starvation tolerance and virulenceof Cryptococcus neoformans.J Clin Invest. 2008 Mar;118(3):1186-97. doi: 10.1172/JCI32053. J Clin Invest. 2008. PMID: 18259613 Free PMC article.
-
Biological functions of the autophagy-related proteins Atg4 and Atg8 in Cryptococcus neoformans.PLoS One. 2020 Apr 6;15(4):e0230981. doi: 10.1371/journal.pone.0230981. eCollection 2020. PLoS One. 2020. PMID: 32251488 Free PMC article.
-
Role of laccase in the biology and virulence of Cryptococcus neoformans.FEMS Yeast Res. 2004 Oct;5(1):1-10. doi: 10.1016/j.femsyr.2004.04.004. FEMS Yeast Res. 2004. PMID: 15381117 Review.
-
Mechanisms of Autophagy.Annu Rev Biophys. 2015;44:101-22. doi: 10.1146/annurev-biophys-060414-034248. Epub 2015 Feb 26. Annu Rev Biophys. 2015. PMID: 25747593 Review.
Cited by
-
Activation of microlipophagy during early infection of insect hosts by Metarhizium robertsii.Autophagy. 2022 Mar;18(3):608-623. doi: 10.1080/15548627.2021.1943179. Epub 2021 Jun 21. Autophagy. 2022. PMID: 34130590 Free PMC article.
-
Mycoplasma bovis inhibits autophagy in bovine mammary epithelial cells via a PTEN/PI3K-Akt-mTOR-dependent pathway.Front Microbiol. 2022 Jul 26;13:935547. doi: 10.3389/fmicb.2022.935547. eCollection 2022. Front Microbiol. 2022. PMID: 35958147 Free PMC article.
-
Autophagy-Related Gene PlATG6a Is Involved in Mycelial Growth, Asexual Reproduction and Tolerance to Salt and Oxidative Stresses in Peronophythora litchii.Int J Mol Sci. 2022 Feb 6;23(3):1839. doi: 10.3390/ijms23031839. Int J Mol Sci. 2022. PMID: 35163762 Free PMC article.
-
Autophagy-related protein PlATG2 regulates the vegetative growth, sporangial cleavage, autophagosome formation, and pathogenicity of peronophythora litchii.Virulence. 2024 Dec;15(1):2322183. doi: 10.1080/21505594.2024.2322183. Epub 2024 Mar 4. Virulence. 2024. PMID: 38438325 Free PMC article.
-
Loss of the putative Rab GTPase, Ypt7, impairs the virulence of Cryptococcus neoformans.Front Microbiol. 2024 Jul 25;15:1437579. doi: 10.3389/fmicb.2024.1437579. eCollection 2024. Front Microbiol. 2024. PMID: 39119141 Free PMC article.
References
LinkOut - more resources
Full Text Sources

