TALEN-based editing of TFIIAy5 changes rice response to Xanthomonas oryzae pv. Oryzae
- PMID: 32029874
- PMCID: PMC7005142
- DOI: 10.1038/s41598-020-59052-w
TALEN-based editing of TFIIAy5 changes rice response to Xanthomonas oryzae pv. Oryzae
Abstract
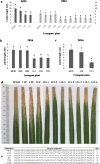
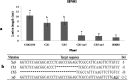
The xa5 gene encodes a basal transcription factor (TFIIAγ) protein with wide spectrum resistance to bacterial blight caused by Xanthomonas oryzae pv. Oryzae (Xoo) in rice. It was only found in a few rice ecotypes, and the recessive characteristics limited its application in breeding. Here, we employed a TALEN-based technique to edit its dominant allelic TFIIAγ5 and obtained many mutant TFIIAγ5 genes. Most of them reduced rice susceptibility to varying degrees when the plants were challenged with the Xoo. In particular, the knocked-out TFIIAγ5 can reduce the rice susceptibility significantly, although it cannot reach the xa5-mediated resistance level, indicating TFIIAγ5 is a major component involved in disease susceptibility. In addition, the mutant encoding the protein with deletion of the 32nd amino acid or amino acid insertion between 32nd and 33rd site confers rice with the similar resistance to that of the knocked-out TFIIAγ5. Thus, the amino acids around 32nd site are also the important action sites of TFIIAγ5 besides the 39th amino acid previously reported. Moreover, the integration of xa5 into TFIIAγ5-knockout plants conferred them with a similar resistance as IRBB5, the rice variety containing the homozygous xa5 gene. Thus, TFIIAγ5 was not simply regarded as a resistant or a susceptible locus, as the substitution of amino acids might shift its functions.
Conflict of interest statement
Dr. Xia’s work has been funded by the National Natural Science Foundation of China (No. 31071173). He has received compensation as a member of Hainan University of China. Dr. Han, Dr. Jiang and Prof. Zhai declare no potential conflict of interest.
Figures



Similar articles
-
Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAgamma1.Mol Genet Genomics. 2006 Apr;275(4):354-66. doi: 10.1007/s00438-005-0091-7. Epub 2006 Jan 28. Mol Genet Genomics. 2006. PMID: 16614777
-
Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice.PLoS Pathog. 2018 Jun 4;14(6):e1007092. doi: 10.1371/journal.ppat.1007092. eCollection 2018 Jun. PLoS Pathog. 2018. PMID: 29864161 Free PMC article.
-
Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae.Plant Biotechnol J. 2015 Sep;13(7):993-1001. doi: 10.1111/pbi.12342. Epub 2015 Feb 3. Plant Biotechnol J. 2015. PMID: 25644581
-
Rice Routes of Countering Xanthomonas oryzae.Int J Mol Sci. 2018 Oct 2;19(10):3008. doi: 10.3390/ijms19103008. Int J Mol Sci. 2018. PMID: 30279356 Free PMC article. Review.
-
Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach.Mol Biol Rep. 2019 Feb;46(1):1519-1532. doi: 10.1007/s11033-019-04584-2. Epub 2019 Jan 9. Mol Biol Rep. 2019. PMID: 30628024 Review.
Cited by
-
Rapid generation of fragrant thermo-sensitive genic male sterile rice with enhanced disease resistance via CRISPR/Cas9.Planta. 2024 Apr 6;259(5):112. doi: 10.1007/s00425-024-04392-4. Planta. 2024. PMID: 38581602
-
Marker-assisted enhancement of bacterial blight (Xanthomonas oryzae pv. oryzae) resistance in a salt-tolerant rice variety for sustaining rice production of tropical islands.Front Plant Sci. 2023 Sep 25;14:1221537. doi: 10.3389/fpls.2023.1221537. eCollection 2023. Front Plant Sci. 2023. PMID: 37818314 Free PMC article.
-
A review of approaches to control bacterial leaf blight in rice.World J Microbiol Biotechnol. 2022 May 17;38(7):113. doi: 10.1007/s11274-022-03298-1. World J Microbiol Biotechnol. 2022. PMID: 35578069 Review.
-
Molecular Basis of Plant-Pathogen Interactions in the Agricultural Context.Biology (Basel). 2024 Jun 6;13(6):421. doi: 10.3390/biology13060421. Biology (Basel). 2024. PMID: 38927301 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

