Gammaherpesvirus-infected germinal center cells express a distinct immunoglobulin repertoire
- PMID: 32029571
- PMCID: PMC7012147
- DOI: 10.26508/lsa.201900526
Gammaherpesvirus-infected germinal center cells express a distinct immunoglobulin repertoire
Abstract
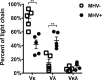
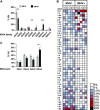
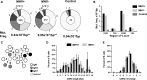
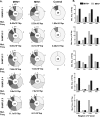
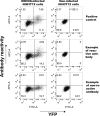
The gammaherpesviruses (γHVs), human Kaposi sarcoma-associated herpesvirus (KSHV), EBV, and murine γHV68 are prevalent infections associated with lymphocyte pathologies. After primary infection, EBV and γHV68 undergo latent expansion in germinal center (GC) B cells and persists in memory cells. The GC reaction evolves and selects antigen-specific B cells for memory development but whether γHV passively transients or manipulates this process in vivo is unknown. Using the γHV68 infection model, we analyzed the Ig repertoire of infected and uninfected GC cells from individual mice. We found that infected cells displayed the hallmarks of affinity maturation, hypermutation, and isotype switching but underwent clonal expansion. Strikingly, infected cells displayed distinct repertoire, not found in uninfected cells, with recurrent utilization of certain Ig heavy V segments including Ighv10-1 In a manner observed with KSHV, γHV68 infected cells also displayed lambda light chain bias. Thus, γHV68 subverts GC selection to expand in a specific B cell subset during the process that develops long-lived immunologic memory.
© 2020 Zelazowska et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

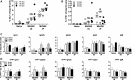




Similar articles
-
Dangerous Liaisons: Gammaherpesvirus Subversion of the Immunoglobulin Repertoire.Viruses. 2020 Jul 23;12(8):788. doi: 10.3390/v12080788. Viruses. 2020. PMID: 32717815 Free PMC article. Review.
-
Murine gammaherpesvirus infection is skewed toward Igλ+ B cells expressing a specific heavy chain V-segment.PLoS Pathog. 2020 Apr 30;16(4):e1008438. doi: 10.1371/journal.ppat.1008438. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32353066 Free PMC article.
-
Role of B-cell proliferation in the establishment of gammaherpesvirus latency.J Virol. 2005 Aug;79(15):9480-91. doi: 10.1128/JVI.79.15.9480-9491.2005. J Virol. 2005. PMID: 16014911 Free PMC article.
-
B Cell-Intrinsic Expression of Interferon Regulatory Factor 1 Supports Chronic Murine Gammaherpesvirus 68 Infection.J Virol. 2020 Jun 16;94(13):e00399-20. doi: 10.1128/JVI.00399-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32321819 Free PMC article.
-
Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner.J Virol. 2007 Jun;81(11):6134-40. doi: 10.1128/JVI.00108-07. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360749 Free PMC article. Review.
Cited by
-
A Mouse Model to Study the Pathogenesis of γ-herpesviral Infections in Germinal Center B Cells.Cells. 2023 Dec 6;12(24):2780. doi: 10.3390/cells12242780. Cells. 2023. PMID: 38132100 Free PMC article.
-
Multifaceted roles for STAT3 in gammaherpesvirus latency revealed through in vivo B cell knockout models.mBio. 2024 Feb 14;15(2):e0299823. doi: 10.1128/mbio.02998-23. Epub 2024 Jan 3. mBio. 2024. PMID: 38170993 Free PMC article.
-
Conquering the Host: Determinants of Pathogenesis Learned from Murine Gammaherpesvirus 68.Annu Rev Virol. 2021 Sep 29;8(1):349-371. doi: 10.1146/annurev-virology-011921-082615. Annu Rev Virol. 2021. PMID: 34586873 Free PMC article.
-
Dangerous Liaisons: Gammaherpesvirus Subversion of the Immunoglobulin Repertoire.Viruses. 2020 Jul 23;12(8):788. doi: 10.3390/v12080788. Viruses. 2020. PMID: 32717815 Free PMC article. Review.
-
Kidins220 regulates the development of B cells bearing the λ light chain.Elife. 2024 Jan 25;13:e83943. doi: 10.7554/eLife.83943. Elife. 2024. PMID: 38271217 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
