A conserved ATG2-GABARAP family interaction is critical for phagophore formation
- PMID: 32009292
- PMCID: PMC7054675
- DOI: 10.15252/embr.201948412
A conserved ATG2-GABARAP family interaction is critical for phagophore formation
Abstract
The intracellular trafficking pathway, macroautophagy, is a recycling and disposal service that can be upregulated during periods of stress to maintain cellular homeostasis. An essential phase is the elongation and closure of the phagophore to seal and isolate unwanted cargo prior to lysosomal degradation. Human ATG2A and ATG2B proteins, through their interaction with WIPI proteins, are thought to be key players during phagophore elongation and closure, but little mechanistic detail is known about their function. We have identified a highly conserved motif driving the interaction between human ATG2 and GABARAP proteins that is in close proximity to the ATG2-WIPI4 interaction site. We show that the ATG2A-GABARAP interaction mutants are unable to form and close phagophores resulting in blocked autophagy, similar to ATG2A/ATG2B double-knockout cells. In contrast, the ATG2A-WIPI4 interaction mutant fully restored phagophore formation and autophagy flux, similar to wild-type ATG2A. Taken together, we provide new mechanistic insights into the requirements for ATG2 function at the phagophore and suggest that an ATG2-GABARAP/GABARAP-L1 interaction is essential for phagophore formation, whereas ATG2-WIPI4 interaction is dispensable.
Keywords: GABARAP; ATG2; autophagosome; autophagy; phagophore.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
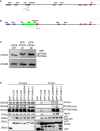
- A, B
Strategy for insertion of GFP‐tag upstream of human ATG2A exon 1. Graphic shows position of guides and locus before and after (B) GFP‐tag plus linker insertion.
- C
Western blot of total cell lysates from parental wild type (WT) and GFP‐ATG2A CRISPR/Cas9 knock‐in clones 1 and 2 using anti‐ATG2A and anti‐ATG2B antibodies.
- D
GFP alone or GFP‐tagged mammalian ATG8 proteins (LC3A, LC3B, LC3C, GABARAP, GABARAP‐L1 and GABARAP‐L2) were overexpressed in HEK293T cells, lysed and the GFP‐tag immunoprecipitated using GFP‐TRAP beads. Samples were then run on 4–12% Bis–Tris gel and transferred to PVDF membrane and blotted for the presence of ATG2A, ATG2B, p62/SQSTM1, WIPI4 and anti‐GFP. Blots are representative of n = 3 independent experiments.
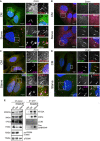
- A
U2OS cells modified to expressed endogenous GFP‐tagged ATG2A (green) were grown in complete media (CM) or starved (EBSS) for 2 h before fixation and immunostaining with antibodies against ATG2B (red) and LC3B (magenta) and analysed by confocal microscopy. Arrows mark ATG2A/LC3B/ATG2B‐positive structures. Scale bar 10 μm.
- B–D
Cells were treated as in (A) and stained with anti‐WIPI2 (red) or (C) anti‐ATG16L1 (red) or (D) anti‐GABARAP‐L1 (red). Arrows mark structures of interest. Scale bar 10 μm.
- E
U2OS WT or U2OS GFP‐ATG2A knock‐in (KI) cells were grown in CM or starvation media for 2 h, lysed and incubated with anti‐GFP nanobody beads coupled to agarose to immunoprecipitate (IP) GFP‐ATG2A. IP samples and 2% input lysates were run on 4–12% gradient gel and processed for Western blotting. Anti‐ATG2A, anti‐WIPI4 and anti‐LC3B, and anti‐GABARAP (pan) were used to probe for the presence/absence of autophagy proteins in the immunoprecipitated samples. p‐p70S6K (T389) was used as a marker for starved cells and total p70S6K as loading control.
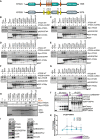
- A
Domain structure of ATG2A (green) and ATG2B (grey) proteins. Both ATG2s contain an N‐terminal VPS13/chorein domain, ATG2 C‐terminal autophagy domain (CAD motif) and ATG2 C‐terminal domain. Position and sequence of putative ATG2 LC3 interaction regions (LIRs) as identified by iLIR and manual annotation. ATG2A has 5 potential LIRs, and ATG2B has 6 potential LIRs. See Table 1 for details.
- B–E
Myc‐tagged ATG2A wild type (WT) and putative LIR mutants, where the potential core motif was mutated to alanine, were used in a pull‐down assay with GST‐tagged mammalian ATG8 proteins. Shown are ATG2‐mLIR#1 (B), ATG2‐mLIR#2 (C), ATG2‐mLIR#3 (D) and ATG2‐mLIR#4 (E). ATG2‐mLIR#5 is shown on (H). ATG2A‐WT or ATG2A‐mLIRs were overexpressed in HEK293T cells, and lysates were incubated with purified GST alone or GST‐tagged LC3A, LC3B, LC3C, GABARAP, GABARAP‐L1 or GABARAP‐L2. Samples were spun, washed and blotted for the presence/absence of Myc‐tagged ATG2A using anti‐Myc antibody. Anti‐p62/SQSTM1 was used as an internal control for the GST pull‐down samples. GST proteins were visualized by Ponceau S staining of membranes.
- F, G
As in (B) but using Myc‐tagged ATG2B‐WT or ATG2B‐mLIR proteins. Shown are ATG2B mLIR#1 (F) and ATG2B mLIR#4 (G). ATG2B‐mLIR#2 was present on an alpha helix, whereas ATG2B‐mLIR#3 and ATG2B‐mLIR#5 were not expressed. Anti‐p62/SQSTM1 was used as an internal control for the GST pull‐down samples. All blots are representative of at least n = 3 independent experiments.
- H
Myc‐tagged ATG2A wild type (WT), ATG2A‐mutant LIR #5 (mLIR; FCIL/AAAA) (upper blots) or Myc‐tagged ATG2B‐WT or ATG2B‐mLIR (FCIL/AAAA; lower blots) were overexpressed in HEK293T cells, and lysates were incubated with purified GST alone or GST‐tagged LC3A, LC3B, LC3C, GABARAP, GABARAP‐L1 or GABARAP‐L2. Samples were spun, washed and blotted for the presence/absence of Myc‐tagged ATG2s using anti‐Myc antibody. GST proteins were visualized by Ponceau S staining of membranes.
- I
U2OS WT and ATG2A/B double‐knockout cell total lysates, analysed for the presence/absence of autophagy marker proteins including ATG2A, ATG2B, p62/SQSTM1, LC3A, LC3B, LC3C, GABARAP‐L1, GABARAP‐L2 and vinculin used as loading control.
- J
GFP alone, GFP‐WIPI4 or GFP‐WIPI4 with increasing concentrations of mCherry‐GABARAP was expressed in HEK293T cells for 24 h, lysed and GFP‐TRAP beads used to immunoprecipitate GFP alone or GFP‐WIPI4. Samples were then probed with antibodies to detect endogenous ATG2A or ATG2B, anti‐GFP and anti‐GABARAP. Blots are representative of n = 3 independent experiments.
- K
Quantification of ATG2A (blue line, round symbols) and ATG2B (grey line and squares) co‐precipitation with GFP‐WIPI4 in the presence of increasing concentrations of mCherry‐GABARAP from (J). Co‐precipitation was normalized to GFP‐WIPI4 alone. Line and error bars are mean ± SD of n = 3 independent experiments.

Domain structure of ATG2A (green) and ATG2B (grey) proteins. Both ATG2s contain an N‐terminal VPS13/chorein domain, ATG2 C‐terminal autophagy domain (CAD motif) and ATG2 C‐terminal domain. Position and sequence of the new ATG2 LIR motif that directs GABARAP interaction (orange) and the previously identified WIPI4 interaction motif (YFS; purple box). Approximately 30 amino acids separate these two motifs in human ATG2A and ATG2B. Multiple sequence alignment of multiple species using Jalview highlights the conservation of these regions. Abbreviations for species: Hs—Homo sapiens; Mm—Mus musculus; Rn—Rattus norvegicus; Pt—Pan‐troglodytes; Xt—Xenopus tropicalis Dr—Danio rerio; Dm—Drosophila melanogaster; Ce—Caenorhabditis elegans; Sp—Schizosaccharomyces pombe; Sc—Saccharomyces cerevisiae.
Myc‐tagged ATG2A wild type (WT) and ATG2A‐mLIR (orange) were co‐expressed with GFP alone, GFP‐LC3B or GFP‐GABARAP in HEK293T cells, lysed and anti‐GFP nanobodies coupled to agarose were used to immunoprecipitate GFP‐tagged proteins. Samples were subjected to Western blotting and probed for the presence of Myc‐ATG2A in immunoprecipitated samples. Anti‐p62/SQSTM1 was used as an internal control for the immunoprecipitated samples.
As in (B) but using Myc‐tagged ATG2B‐WT or ATG2B‐mLIR (orange) co‐expressed with GFP alone, GFP‐LC3B or GFP‐GABARAP. Samples were subjected to Western blotting and probed for the presence of Myc‐ATG2B in immunoprecipitated samples. Anti‐p62/SQSTM1 was used as an internal control for the immunoprecipitated samples.
HA‐tagged ATG2A‐WT, ATG2A‐mLIR (orange) and ATG2A‐mYFS (YFS/AAA; purple) were stably expressed in U2OS ATG2A/B double‐knockout cells using retrovirus transduction. Cells were transfected with GFP‐GABARAP and 24 h later grown in complete medium (CM) or starved for 2 h (EBSS). Cells were lysed, and anti‐HA beads were used to immunoprecipitate HA‐tagged ATG2A and processed for Western blot. Blots were then probed with antibodies against HA‐tag (ATG2A), anti‐WIPI4 and anti‐GFP for the presence/absence in immunoprecipitated samples. All blots are representative of at least n = 3 independent experiments.
Quantification of WIPI4 and GFP‐GABARAP‐ii co‐precipitation with HA‐ATG2A from (D). Bands were normalized against HA‐ATG2A immunoprecipitate and expressed as a fold change compared with HA‐ATG2A‐WT (complete media; CM). Each symbol represents an independent experiment (n = 3) and error bars mean ± SD.
ATG2A/B double‐knockout cells (DKO) stably expressing HA‐tagged ATG2A‐WT, ATG2A‐mLIR (orange) or ATG2A‐mYFS (YFS/AAA; purple) were transfected with GFP‐GABARAP and 24 h later grown in complete medium (CM) or starved for 2 h (EBSS). Cells were lysed, and GFP‐GABARAP was immunoprecipitated using anti‐GFP‐TRAP beads. Samples were processed for Western blot and probed with anti‐HA‐tag (ATG2A), anti‐WIPI4 and anti‐GFP for their presence/absence in immunoprecipitated samples. All blots are representative of at least n = 3 independent experiments.
Quantification of HA‐ATG2A and WIPI4 co‐precipitation with GFP‐GABARAP from (F). Results were expressed as a fold change compared with HA‐ATG2A‐WT (complete media; CM). Each symbol represents an independent experiment (n = 3) with error bars mean ± SD.
GFP alone, GFP‐GABARAP or GFP‐GABARAP with increasing concentrations of mCherry‐WIPI4 was expressed in HEK293T cells for 24 h, lysed and GFP‐TRAP beads used to immunoprecipitate GFP alone or GFP‐GABARAP. Samples were then probed with antibodies to detect endogenous ATG2A or ATG2B in immunoprecipitated samples. Blots are representative of n = 3 independent experiments.
Quantification of ATG2A (green line, round symbols) and ATG2B (grey line and squares) co‐precipitation with GFP‐GABARAP in the presence of increasing concentrations of mCherry‐WIPI4 from (H). Co‐precipitation was normalized to GFP‐GABARAP alone. Line and error bars are mean ± SD of n = 3 independent experiments.
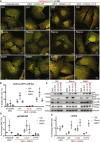
- A
U2OS ATG2A/B double‐knockout (DKO) CRISPR/Cas9 cells stably expressing tandem‐tagged LC3B (mCherry‐GFP‐LC3B) were retrovirally transduced to express vector, or HA‐tagged ATG2A‐WT, ATG2A‐mLIR (FCIL/AAAA) or ATG2A‐mYFS (YFS/AAA). Cells were grown in complete medium (CM) or starved for 2 h (EBSS) or treated with CM plus bafilomycin A1 (200 nM, 4 h), fixed and analysed by confocal microscopy. Merged images of GFP (green) and mCherry (red) channels show the presence of autophagosomes/phagophores (GFP‐ and mCherry‐positive, yellow puncta) or autolysosomes (mCherry only, red puncta) Scale bar 10 μm. Images are representative of n = 3 independent experiments.
- B
Quantification of (A) using flow cytometry of measuring GFP and mCherry fluorescence. Cells were gated based on GFP and mCherry fluorescence and % mCherry‐positive cells gated used as an indication of autolysosome formation due to GFP quenching. Each symbol represents 1 independent experiment with 10,000 cells analysed per condition. A total of n = 3 independent experiments were performed, and horizontal bar indicates mean ± SD.
- C
U2OS ATG2A/B DKO cells reconstituted with vector only, HA‐ATG2A‐WT, HA‐ATG2A‐mLIR and HA‐ATG2A‐mYFS were stimulated with complete medium (CM), 2‐h starvation (EBSS) or 4‐h bafilomycin A1 (BafA1, 200 nM), lysed in total cell lysis buffer and subjected to Western blot analysis. Blots were probed for the presence of HA‐tag (ATG2A), p62/SQSTM1, LC3B, pan‐GABARAP (GABARAP, GABARAP‐L1 and GABARAP‐L2) and vinculin (loading control).
- D, E
p62/SQSTM1 (D) and LC3B‐II (E) levels were normalized to loading control and quantified as fold change of DKO proteins levels. Each symbol represents an independent experiment. Quantification of at least n = 3 independent experiments is shown. Horizontal bar represents mean ± SD.
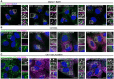
- A
U2OS ATG2A/B DKO cells reconstituted with vector only, HA‐ATG2A‐WT, HA‐ATG2A‐mLIR and HA‐ATG2A‐mYFS were stimulated starvation (EBSS) plus bafilomycin A1 (BafA1, 200 nM) for 4 h to stimulate autophagosome generation and prevent their degradation in the lysosome. Cells were fixed and immune‐stained for LC3B (green) and LAMP2 (magenta) to visualize lysosomes. DAPI was included (blue) to mark the DNA/nucleus. Closed arrows (ATG2A/B DKO and DKO + ATG2A mLIR) highlight aggregate structures. Open arrows (DKO + ATG2A‐WT and ATG2A‐mYFS) highlight LAMP2/LC3B‐positive vesicles. Scale bar 10 μm.
- B, C
U2OS ATG2A/B DKO cells reconstituted with vector only, HA‐ATG2A‐WT, HA‐ATG2A‐mLIR and HA‐ATG2A‐mYFS were stimulated with either 2% BSA only (B) or 2% BSA plus 500 μM oleic acid (C) for 16 h prior to fixation in 4% PFA. Cells were permeabilized using saponin and stained with anti‐HA (ATG2A; magenta) and 5 μM BODIPY 493/503 to visualize lipid droplets (green; LDs). DAPI was included (blue) to mark the DNA/nucleus. Open arrows highlight ATG2A‐positive lipid droplets. All images are representative of at least n = 3 independent experiments. Scale bar 10 μm.
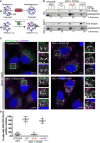
Graphical representation of proteinase K assay (i) showing protection of p62/SQSTM1 inside sealed autophagosomes or proteinase K‐sensitive p62 present within open phagophores. (ii) Graphical representation of sytnatxin17 (STX17) translocation to completed autophagosomes and not phagophores. Autophagosomes are identified as being both LC3B‐ and STX17‐positive vesicles.
U2OS ATG2A/B DKO cells reconstituted with vector only, HA‐ATG2A‐WT, HA‐ATG2A‐mLIR and HA‐ATG2A‐mYFS were stimulated with complete medium (CM), 4‐h starvation (EBSS) plus bafilomycin A1 (BafA1, 200 nM) treatment. Cells were centrifuged and resuspended in PBS digitonin, spun, washed and the membrane fractions incubated with proteinase K with and without 0.1% Triton X‐100. Samples were then subjected to Western blotting using anti‐p62/SQSTM1 and anti‐HA (ATG2A) antibodies. Percentage p62/SQSTM1 remaining was calculated using densitometry analysis. Blots are representative of n = 3 independent experiments.
U2OS ATG2A/B DKO cells reconstituted with vector only, HA‐ATG2A‐WT, HA‐ATG2A‐mLIR and HA‐ATG2A‐mYFS and stably expressing GFP‐Syntaxin 17 (STX17) were stimulated starvation (EBSS) plus bafilomycin A1 (BafA1, 200 nM) for 4 h to stimulate autophagosome generation and prevent their degradation in the lysosome. Cells were fixed and immune‐stained for LC3B (magenta). DAPI was included (blue) to mark the DNA/nucleus. Confocal analysis of LC3B and GFP‐STX17 (green) localization was performed. Closed arrows (ATG2A/B DKO and DKO + ATG2A mLIR) highlight aggregate structures. Open arrows (DKO + ATG2A‐WT and ATG2A‐mYFS) highlight STX17/LC3B‐positive vesicles. Scale bar 10 μm.
Quantification of (C) expressed as a percentage of cells with STX17/LC3B‐positive vesicles. Each symbol represents a single field of cells with 5–10 cells per field. A total of 300–600 cells were analysed over n = 3 independent experiments. Data are shown as mean ± SD.
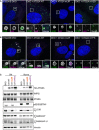
- A, B
U2OS ATG2A/B DKO cells reconstituted with vector only, HA‐ATG2A‐WT, HA‐ATG2A‐mLIR and HA‐ATG2A‐mYFS were stimulated starvation (EBSS) 2 h to stimulate autophagosome generation. Cells were then fixed and immune‐stained for LC3B (green), p62/SQSTM1 (magenta) (A) or GABARAP‐L1 (magenta) (B) and DAPI (blue; DNA/nucleus). Images were taken on a Zeiss 880 AiryScan super‐resolution confocal microscope. All images are representative of at least n = 3 independent experiments. Scale bar 10 μm.
- C
Total cell lysates of ATG2A/B double‐knockout cells alone or reconstituted with HA‐tagged ATG2A‐WT, ATG2A‐mLIR (FCIL/AAAA) or ATG2A‐mYFS (YFS/AAA) and left in complete media (CM) or starved for 2 h (EBSS). Cells were lysed and blotted for autophagy marker proteins p62/SQSTM1, LC3B, ATG9A, WIPI2, GABARAP and GABARAP‐L1. Anti‐HA for ATG2A expression and vinculin were used as a loading control.
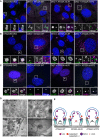
- A–C
U2OS ATG2A/B double‐knockout (DKO) cells or DKO reconstituted with ATG2A‐WT, ATG2A‐mLIR (FCIL/AAAA) or ATG2A‐mYFS (YFS/AAA) were starved for 2 h, fixed and stained for LC3B (green) or WIPI2 (magenta), (B) ATG9A (magenta) or (C) GABARAP (Magenta) and imaged using a Zeiss 880 AiryScan super‐resolution confocal microscope. Images are representatives of n = 3 independent experiments. Scale bar 10 μm.
- D
Transmission electron micrographs of ATG2A/B DKO cells (upper left), DKO + ATG2A‐WT (upper right), DKO+ ATG2A‐mLIR (lower left) and DKO+ATG2A‐mYFS (lower right) starved (EBSS) for 2 h. Clustered small vesicles (open arrowheads; DKO and DKO‐mLIR) and autophagosomes (closed arrow heads, DKO+ATG2A‐WT and DKO+ATG2A‐mYFS) indicated. Images are representative of n = 3 independent experiments. Scale bar 500 nm. ER = endoplasmic reticulum; N = nucleus; Mt = mitochondria; G = Golgi; AP = autophagosome.
- E
Model of ATG2 function based on the current knowledge. ATG2A localizes to ER membranes and facilitates lipid transfer from the ER to the growing phagophore. ATG2 interaction with GABARAP/GABARAP‐L1 is essential for anchoring ATG2 to growing phagophore and mutation of the GABARAP interaction region results in the formation of immature phagophores.
Similar articles
-
Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex.Proc Natl Acad Sci U S A. 2018 Oct 16;115(42):E9792-E9801. doi: 10.1073/pnas.1811874115. Epub 2018 Sep 5. Proc Natl Acad Sci U S A. 2018. PMID: 30185561 Free PMC article.
-
TOM40 Targets Atg2 to Mitochondria-Associated ER Membranes for Phagophore Expansion.Cell Rep. 2019 Aug 13;28(7):1744-1757.e5. doi: 10.1016/j.celrep.2019.07.036. Cell Rep. 2019. PMID: 31412244 Free PMC article.
-
ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system.Mol Cell. 2021 May 6;81(9):2013-2030.e9. doi: 10.1016/j.molcel.2021.03.001. Epub 2021 Mar 26. Mol Cell. 2021. PMID: 33773106
-
Mechanistic Insights into the Role of Atg11 in Selective Autophagy.J Mol Biol. 2020 Jan 3;432(1):104-122. doi: 10.1016/j.jmb.2019.06.017. Epub 2019 Jun 22. J Mol Biol. 2020. PMID: 31238043 Free PMC article. Review.
-
Lipids and membrane-associated proteins in autophagy.Protein Cell. 2021 Jul;12(7):520-544. doi: 10.1007/s13238-020-00793-9. Epub 2020 Nov 5. Protein Cell. 2021. PMID: 33151516 Free PMC article. Review.
Cited by
-
Sequence Analysis and Structural Predictions of Lipid Transfer Bridges in the Repeating Beta Groove (RBG) Superfamily Reveal Past and Present Domain Variations Affecting Form, Function and Interactions of VPS13, ATG2, SHIP164, Hobbit and Tweek.Contact (Thousand Oaks). 2022 Jan;5:251525642211343. doi: 10.1177/25152564221134328. Epub 2022 Nov 21. Contact (Thousand Oaks). 2022. PMID: 36571082 Free PMC article.
-
Loss of Profilin3 Impairs Spermiogenesis by Affecting Acrosome Biogenesis, Autophagy, Manchette Development and Mitochondrial Organization.Front Cell Dev Biol. 2021 Nov 4;9:749559. doi: 10.3389/fcell.2021.749559. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34869336 Free PMC article.
-
The transcription factor NRF1 (NFE2L1) activates aggrephagy by inducing p62 and GABARAPL1 after proteasome inhibition to maintain proteostasis.Sci Rep. 2023 Sep 1;13(1):14405. doi: 10.1038/s41598-023-41492-9. Sci Rep. 2023. PMID: 37658135 Free PMC article.
-
Ciliary Type III Adenylyl Cyclase in the VMH Is Crucial for High-Fat Diet-Induced Obesity Mediated by Autophagy.Adv Sci (Weinh). 2022 Jan;9(3):e2102568. doi: 10.1002/advs.202102568. Epub 2021 Nov 16. Adv Sci (Weinh). 2022. PMID: 34783461 Free PMC article.
-
The Cross-Links of Endoplasmic Reticulum Stress, Autophagy, and Neurodegeneration in Parkinson's Disease.Front Aging Neurosci. 2021 Jun 3;13:691881. doi: 10.3389/fnagi.2021.691881. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34168552 Free PMC article. Review.
References
-
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P et al (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

