Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study
- PMID: 32004165
- PMCID: PMC7147275
- DOI: 10.1097/CM9.0000000000000722
Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study
Abstract
Background: Human infections with zoonotic coronaviruses (CoVs), including severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV, have raised great public health concern globally. Here, we report a novel bat-origin CoV causing severe and fatal pneumonia in humans.
Methods: We collected clinical data and bronchoalveolar lavage (BAL) specimens from five patients with severe pneumonia from Wuhan Jinyintan Hospital, Hubei province, China. Nucleic acids of the BAL were extracted and subjected to next-generation sequencing. Virus isolation was carried out, and maximum-likelihood phylogenetic trees were constructed.
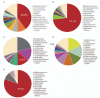
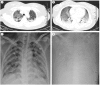
Results: Five patients hospitalized from December 18 to December 29, 2019 presented with fever, cough, and dyspnea accompanied by complications of acute respiratory distress syndrome. Chest radiography revealed diffuse opacities and consolidation. One of these patients died. Sequence results revealed the presence of a previously unknown β-CoV strain in all five patients, with 99.8% to 99.9% nucleotide identities among the isolates. These isolates showed 79.0% nucleotide identity with the sequence of SARS-CoV (GenBank NC_004718) and 51.8% identity with the sequence of MERS-CoV (GenBank NC_019843). The virus is phylogenetically closest to a bat SARS-like CoV (SL-ZC45, GenBank MG772933) with 87.6% to 87.7% nucleotide identity, but is in a separate clade. Moreover, these viruses have a single intact open reading frame gene 8, as a further indicator of bat-origin CoVs. However, the amino acid sequence of the tentative receptor-binding domain resembles that of SARS-CoV, indicating that these viruses might use the same receptor.
Conclusion: A novel bat-borne CoV was identified that is associated with severe and fatal respiratory disease in humans.
Conflict of interest statement
None.
Figures


Similar articles
-
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding.Lancet. 2020 Feb 22;395(10224):565-574. doi: 10.1016/S0140-6736(20)30251-8. Epub 2020 Jan 30. Lancet. 2020. PMID: 32007145 Free PMC article.
-
Epidemiological, clinical, and virological characteristics of 465 hospitalized cases of coronavirus disease 2019 (COVID-19) from Zhejiang province in China.Influenza Other Respir Viruses. 2020 Sep;14(5):564-574. doi: 10.1111/irv.12758. Epub 2020 May 19. Influenza Other Respir Viruses. 2020. PMID: 32397011 Free PMC article.
-
A COVID-19 patient with multiple negative results for PCR assays outside Wuhan, China: a case report.BMC Infect Dis. 2020 Jul 16;20(1):517. doi: 10.1186/s12879-020-05245-7. BMC Infect Dis. 2020. PMID: 32677909 Free PMC article.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
Emergence of a Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2: Biology and Therapeutic Options.J Clin Microbiol. 2020 Apr 23;58(5):e00187-20. doi: 10.1128/JCM.00187-20. Print 2020 Apr 23. J Clin Microbiol. 2020. PMID: 32161092 Free PMC article. Review.
Cited by
-
Risk Factors Associated with Prolonged Nasopharyngeal Carriage of SARS-CoV-2 and Length of Stay among Patients Admitted to a COVID-19 Referral Center in Manila, Philippines.Acta Med Philipp. 2023 Dec 18;57(12):66-72. doi: 10.47895/amp.vi0.5764. eCollection 2023. Acta Med Philipp. 2023. PMID: 39429759 Free PMC article.
-
Computational Investigations to Identify Potent Natural Flavonoid Inhibitors of the Nonstructural Protein (NSP) 16/10 Complex Against Coronavirus.Cureus. 2024 Aug 29;16(8):e68098. doi: 10.7759/cureus.68098. eCollection 2024 Aug. Cureus. 2024. PMID: 39347210 Free PMC article.
-
Machine learning predictive modeling of the persistence of post-Covid19 disorders: Loss of smell and taste as case studies.Heliyon. 2024 Jul 27;10(15):e35246. doi: 10.1016/j.heliyon.2024.e35246. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170549 Free PMC article.
-
Evaluation of SARS-CoV-2-Specific IgY Antibodies: Production, Reactivity, and Neutralizing Capability against Virus Variants.Int J Mol Sci. 2024 Jul 21;25(14):7976. doi: 10.3390/ijms25147976. Int J Mol Sci. 2024. PMID: 39063218 Free PMC article.
-
Development and characterization of a multimeric recombinant protein using the spike protein receptor binding domain as an antigen to induce SARS-CoV-2 neutralization.Immun Inflamm Dis. 2024 Jul;12(7):e1353. doi: 10.1002/iid3.1353. Immun Inflamm Dis. 2024. PMID: 39056544 Free PMC article.
References
-
- Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 2009; 234:1117–1127. doi: 10.3181/0903-MR-94. - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–1820. doi: 10.1056/NEJMoa1211721. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

